Search Count: 222
 |
Cryo-Em Structure Of Violaxanthin-Chlorophyll-A-Binding Protein With Red Shifted Chl A (Rvcp) From Trachydiscus Minutus At 2.4 Angstrom
Organism: Trachydiscus minutus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, A1L1F, XAT |
 |
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: HYDROLASE Ligands: GOL, CA, TLA |
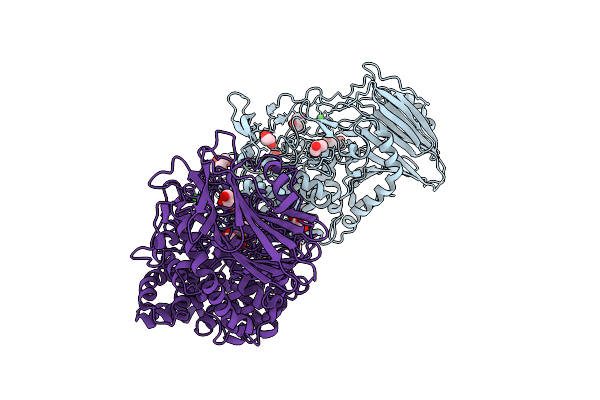 |
Crystal Structure Of Tegh116 From Thermosynechococcus Elongatus With Glucose
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: HYDROLASE Ligands: BGC, GOL, CA, TLA |
 |
Organism: Mus musculus, Chikungunya virus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Mus musculus, Chikungunya virus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
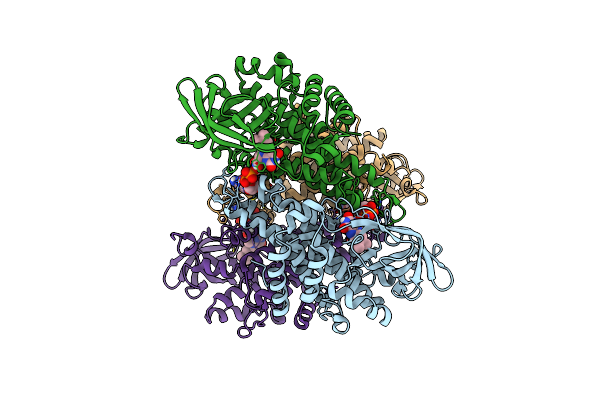
X-Ray Structure Of Cytochrome P450 Olet From Lacicoccus Alkaliphilus In Complex With Icosanoic Acid
Organism: Lacicoccus alkaliphilus dsm 16010
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: HEM, DCR, GOL |
 |
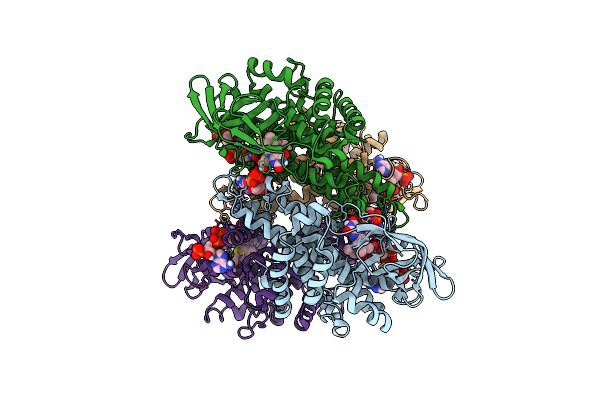
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: HYDROLASE Ligands: FAD |
 |
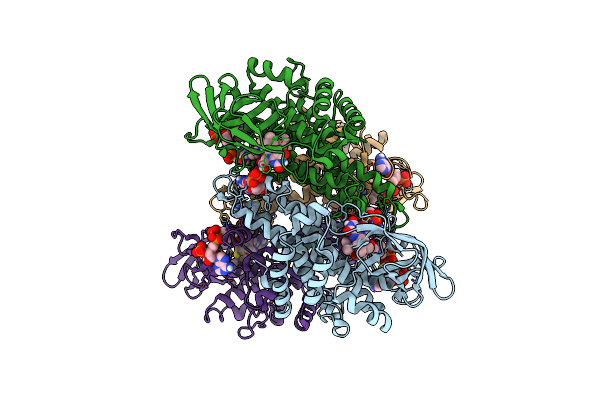
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: HYDROLASE Ligands: BCO, FAD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: HYDROLASE Ligands: IVC, FAD |
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Manganese (Mn) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Iron (Fe) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Zinc (Zn) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of A Cupin Protein (Tm1459, C106V Mutant) In Iron (Fe) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1S |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1R |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1S |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: HXC, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: CO8, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: CAA, MG |

