Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
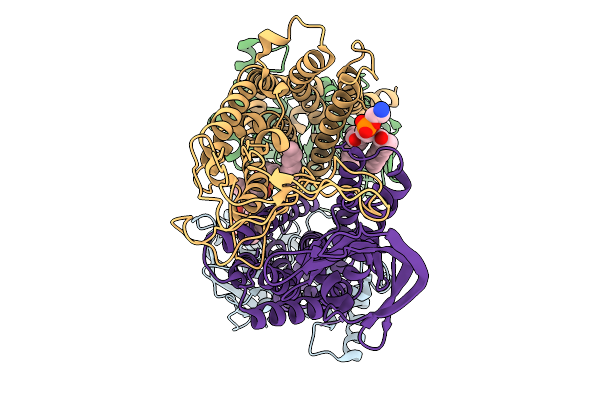 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
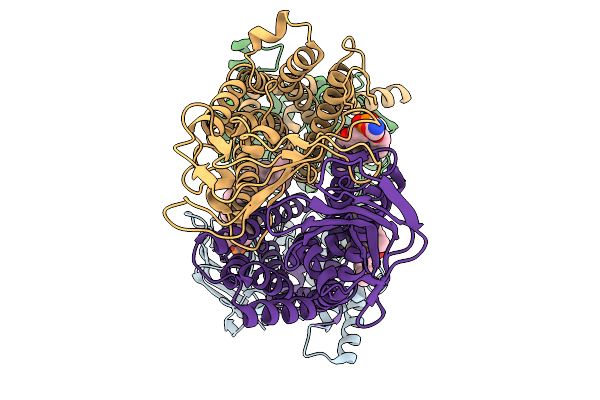 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
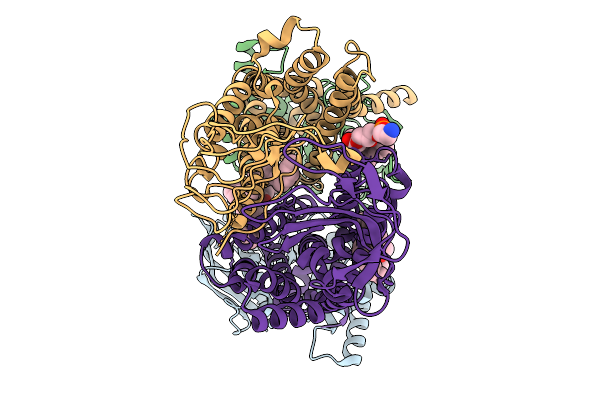 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
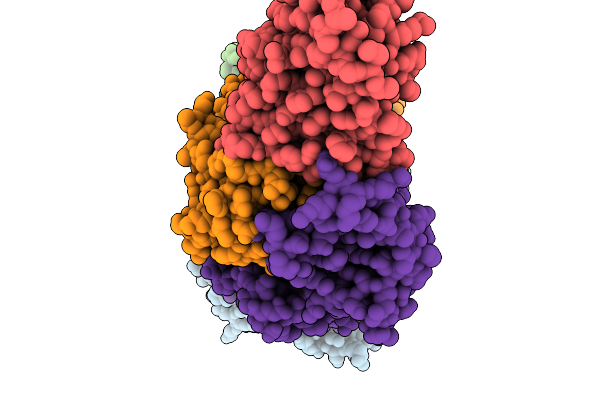 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT |
 |
Integrin Aiibb3 Bent Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
 |
Integrin Aiibb3 Intermediate Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
 |
Integrin Aiibb3 Dimer Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: K, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: K, Y7Z, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:3.62 Å Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-03-19 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-03-19 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-03-19 Classification: TRANSFERASE Ligands: ZN, SR |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSLOCASE Ligands: A1AN8 |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSLOCASE Ligands: A1AON |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSLOCASE Ligands: 3PE, A1AOE |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSLOCASE Ligands: 3PE, A1AOF |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: CYTOSOLIC PROTEIN |