Search Count: 380
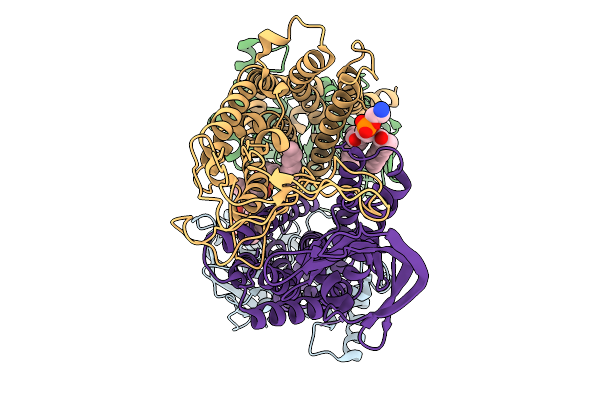 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
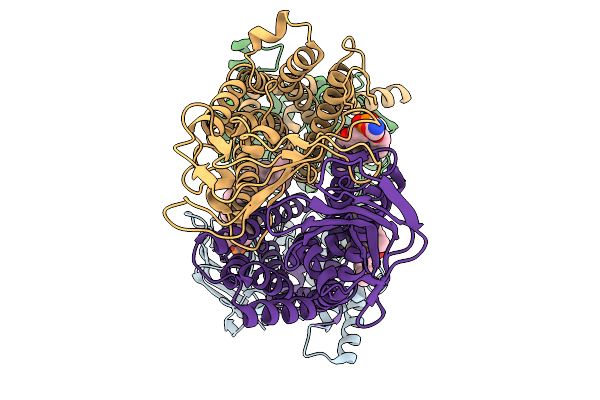 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
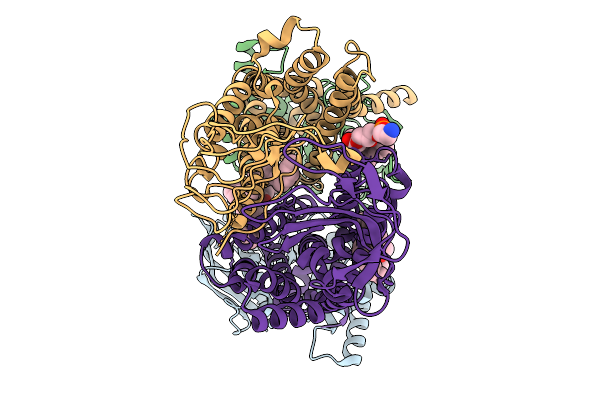 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
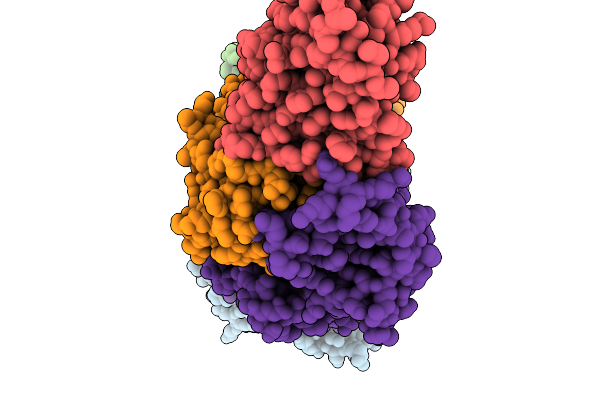 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT |
 |
Cryo-Em Structure Of Human Histamine Receptor H4R In Complex With Agonist Histamine And Gi Proteins
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CLR, HSM |
 |
Integrin Aiibb3 Bent Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
 |
Integrin Aiibb3 Intermediate Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
 |
Integrin Aiibb3 Dimer Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
 |
Crystal Structure Of Ddb1-Crbn-Alv1 Complex Bound To Triple Znf Of Helios (Ikzf2 Zf1-3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.11 Å Release Date: 2025-09-03 Classification: LIGASE Ligands: RN9, ZN |
 |
Cryo-Em Structure Of Mjhku4R-Cov-1 Receptor-Binding Domain Complexed With Human Cd26
Organism: Homo sapiens, Tylonycteris bat coronavirus hku4
Method: ELECTRON MICROSCOPY Resolution:2.61 Å Release Date: 2025-09-03 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Organism: Mirabilis jalapa
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: CIT, FE2, TRS, GOL, PEG |
 |
Crystal Structure Of 1-2C-T96F Tcr In Complex With Hla-A*11:01 Bound To Kras-G12V Peptide (Vvgavgvgk)
Organism: Homo sapiens, Mus musculus, Kirsten murine sarcoma virus
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2025-06-18 Classification: IMMUNE SYSTEM |
 |
Organism: Sfts virus js4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-06-18 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Sfts virus hb29, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-06-18 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe fever with thrombocytopenia syndrome virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2025-06-18 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: K, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: K, Y7Z, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:3.62 Å Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: K, CA |

