Search Count: 25
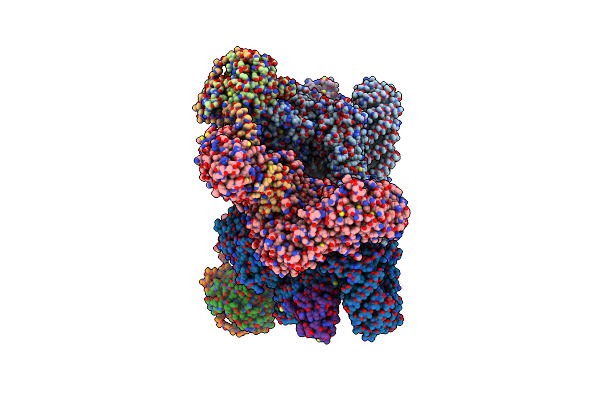 |
Cryo-Em Structure Of Deptor Bound To Human Mtor Complex 2, Overall Refinement
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-15 Classification: SIGNALING PROTEIN Ligands: IHP, ZN, ACE |
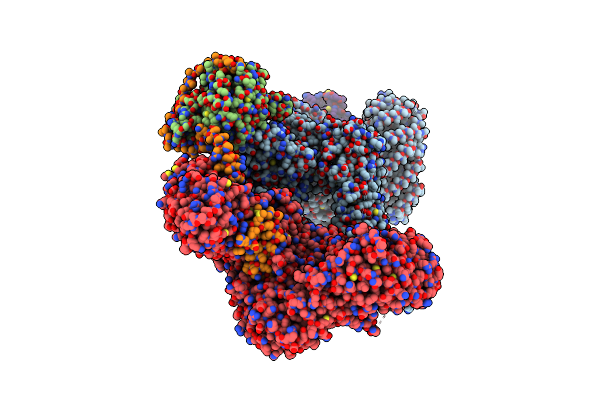 |
Cryo-Em Structure Of Deptor Bound To Human Mtor Complex 2, Focussed On One Protomer
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: IHP, ZN, ACE |
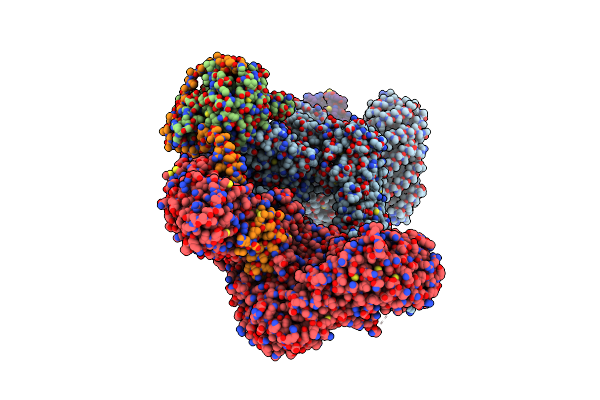 |
Cryo-Em Structure Of Deptor Bound To Human Mtor Complex 2, Dept-Bound Subset Local Refinement
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: IHP, ZN, ACE |
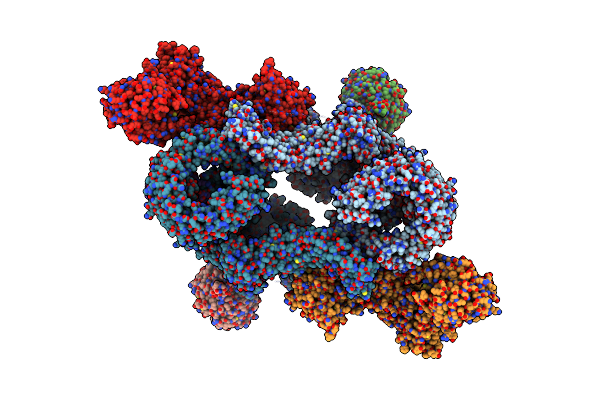 |
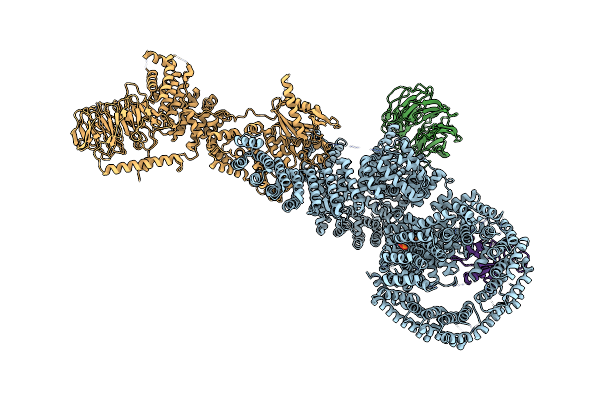
Cryo-Em Structure Of Deptor Bound To Human Mtor Complex 1, Overall Refinement
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: IHP |
 |
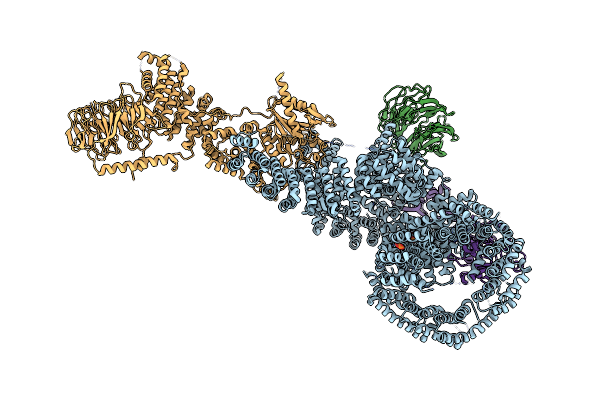
Cryo-Em Structure Of Deptor Bound To Human Mtor Complex 1, Focussed On One Protomer
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: IHP |
 |
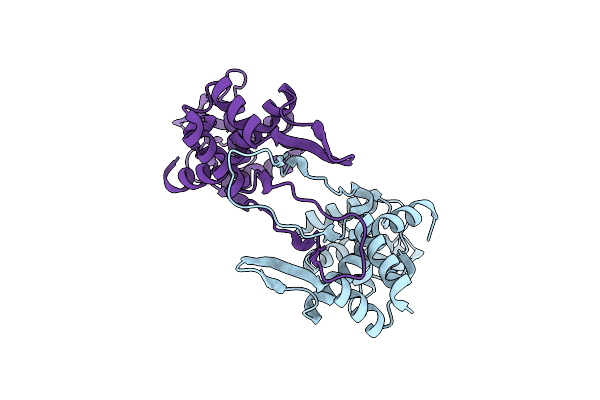
Cryo-Em Structure Of Deptor Bound To Human Mtor Complex 1, Dept-Bound Subset Local Refinement
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: IHP |
 |
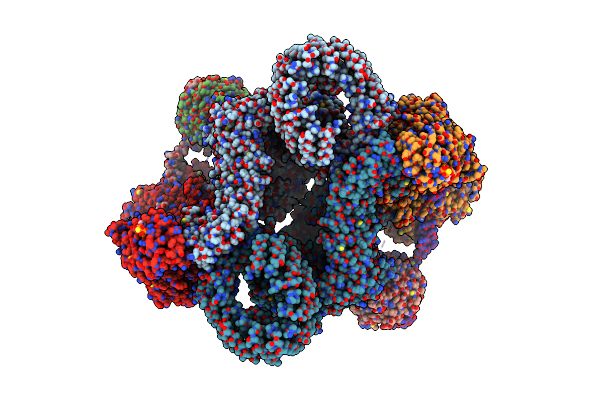
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-09-08 Classification: SIGNALING PROTEIN |
 |
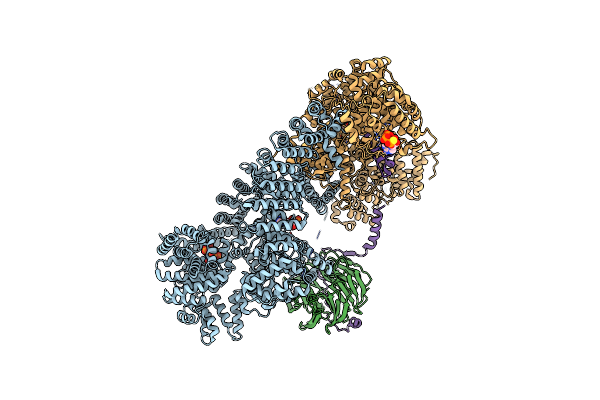
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: AGS, IHP, ZN, ACE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: AGS, IHP, ZN, ACE |
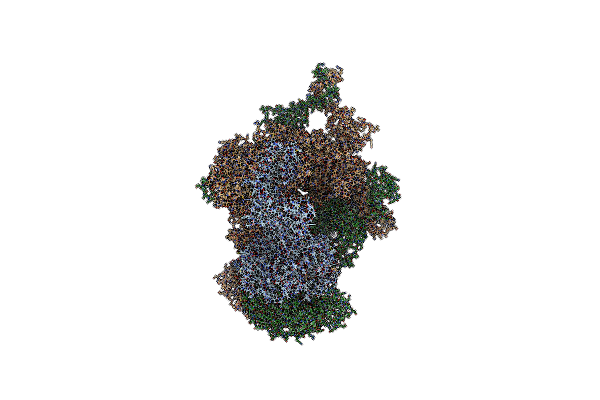 |
Citrate-Induced Acetyl-Coa Carboxylase (Acc-Cit) Filament At 5.4 A Resolution
|
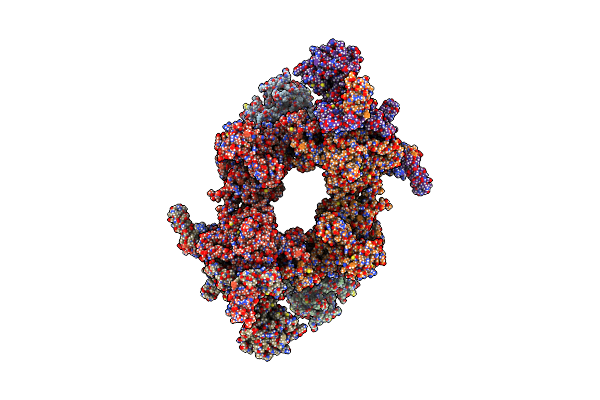 |
Filament Of Acetyl-Coa Carboxylase And Brct Domains Of Brca1 (Acc-Brct) Core At 4.6 A Resolution
|
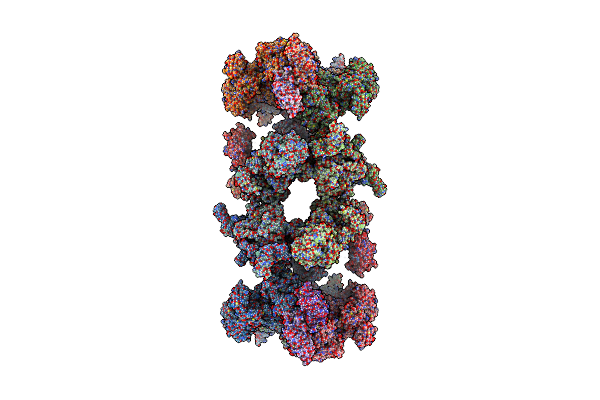 |
Filament Of Acetyl-Coa Carboxylase And Brct Domains Of Brca1 (Acc-Brct) At 5.9 A Resolution
|
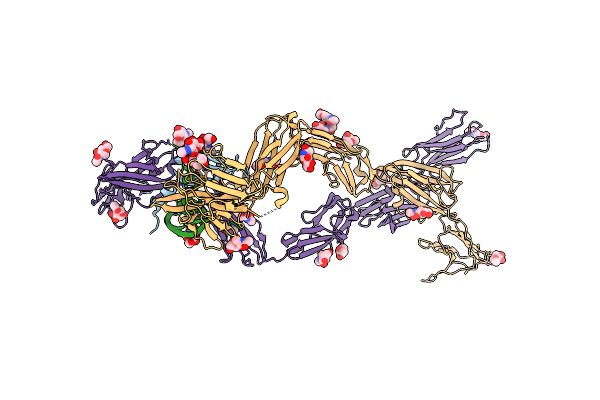 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2017-02-01 Classification: TRANSFERASE Ligands: NAG |
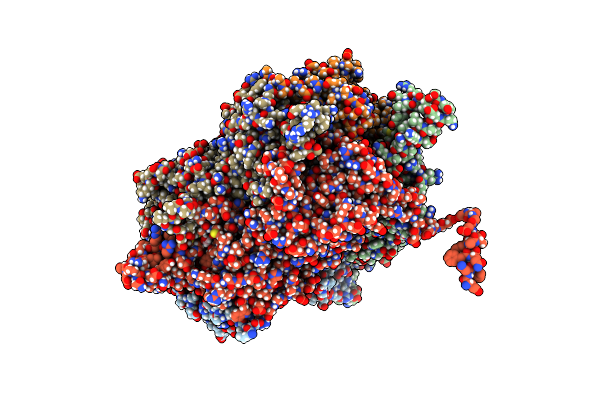 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2016-07-20 Classification: LIGASE Ligands: EDO |
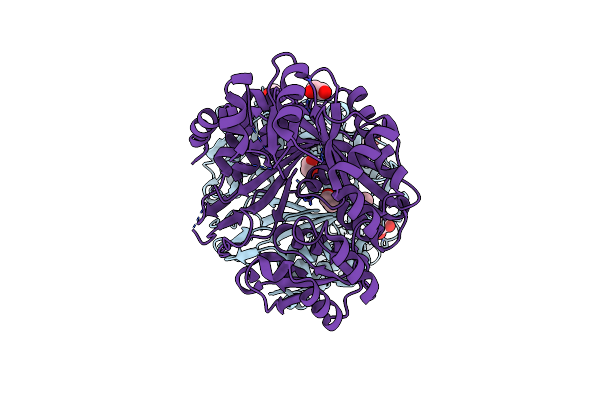 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-07-20 Classification: LIGASE Ligands: EDO |
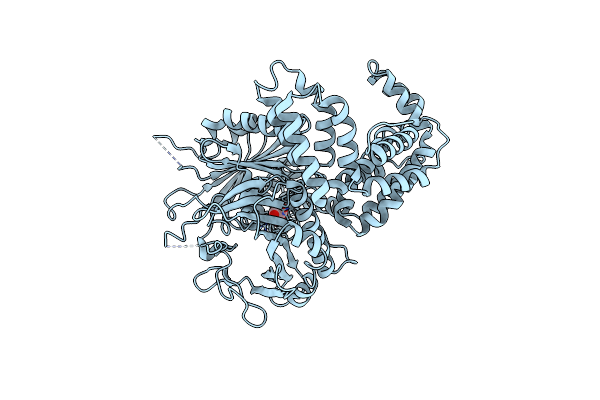 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-04-20 Classification: LIGASE Ligands: MLI |
 |
Crystal Structure Of C-Terminal Variant 1 Of Chaetomium Thermophilum Acetyl-Coa Carboxylase
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2016-04-20 Classification: LIGASE |
 |
Crystal Structure Of C-Terminal Variant 2 Of Chaetomium Thermophilum Acetyl-Coa Carboxylase
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Chaetomium thermophilum var. thermophilum dsm 1495
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2016-04-20 Classification: LIGASE |
 |
Crystal Structure Of Cd-Ct Domains Of Chaetomium Thermophilum Acetyl-Coa Carboxylase
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Chaetomium thermophilum var. thermophilum dsm 1495
Method: X-RAY DIFFRACTION Resolution:7.20 Å Release Date: 2016-04-20 Classification: LIGASE |
 |
Crystal Structure Of A Dbccp-Variant Of Chaetomium Thermophilum Acetyl-Coa Carboxylase
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Chaetomium thermophilum var. thermophilum dsm 1495
Method: X-RAY DIFFRACTION Resolution:8.40 Å Release Date: 2016-04-20 Classification: LIGASE |

