Search Count: 19
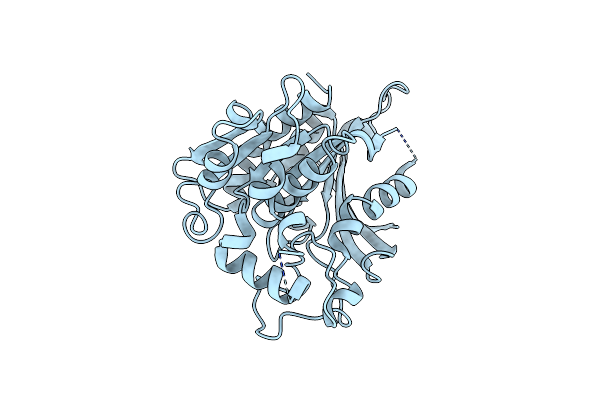 |
Reaction Product Of Paraoxon And S-Formylglutathione Hydrolase W197I Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2008-08-12 Classification: HYDROLASE |
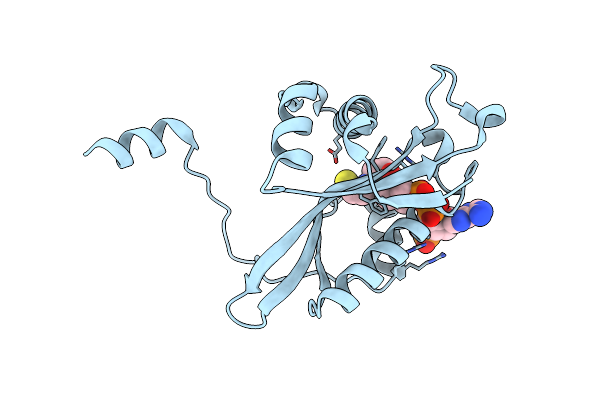 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-01-24 Classification: TRANSFERASE Ligands: COA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-01-17 Classification: TRANSFERASE Ligands: SO4 |
 |
Spermine Spermidine Acetyltransferase In Complex With Acetylcoa, K26R Mutant
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-01-17 Classification: TRANSFERASE Ligands: ACO |
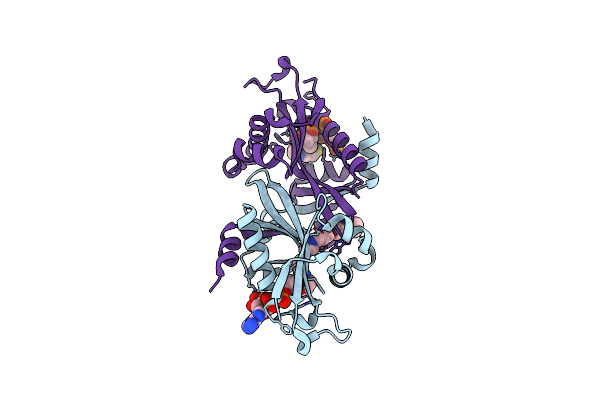 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-01-17 Classification: TRANSFERASE Ligands: COA, B33 |
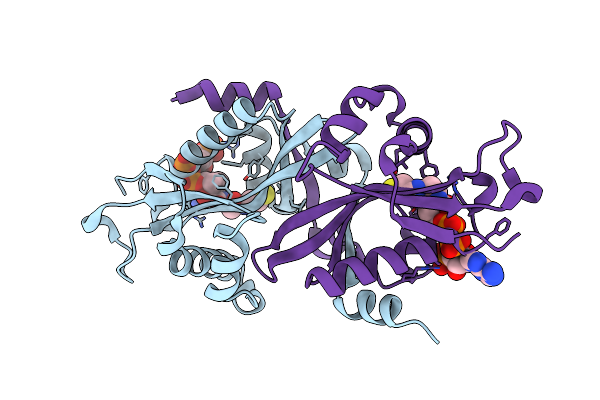 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-01-17 Classification: TRANSFERASE Ligands: COA |
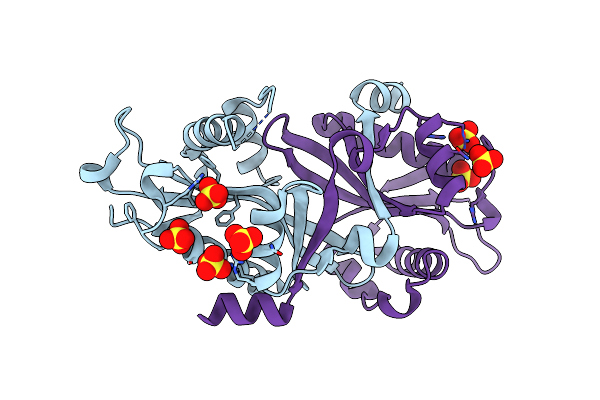 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-01-17 Classification: TRANSFERASE Ligands: SO4 |
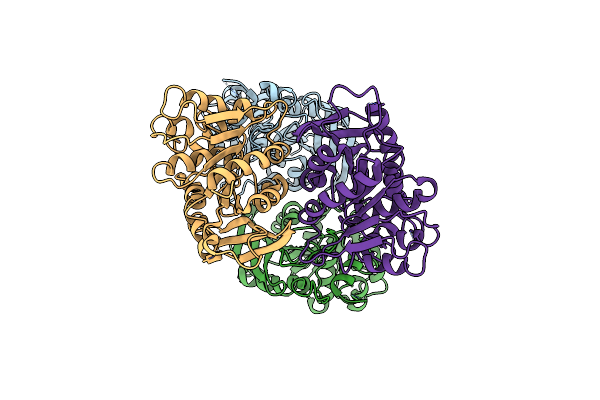 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-11-30 Classification: HYDROLASE |
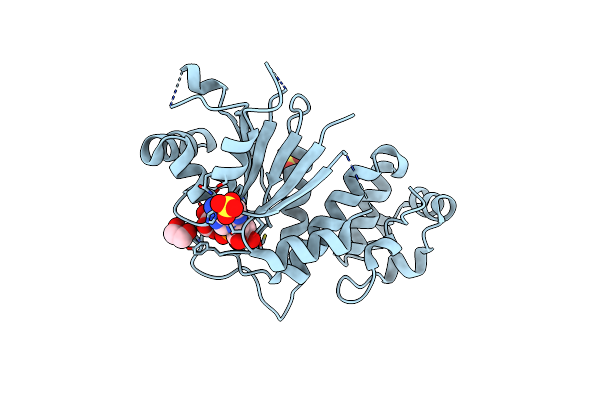 |
Crystal Structure Of Yeast Ymx7, An Adp-Ribose-1''-Monophosphatase, Complexed With Adp-Ribose
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2004-11-30 Classification: structural genomics, unknown function Ligands: SO4, NA, APR, EDO |
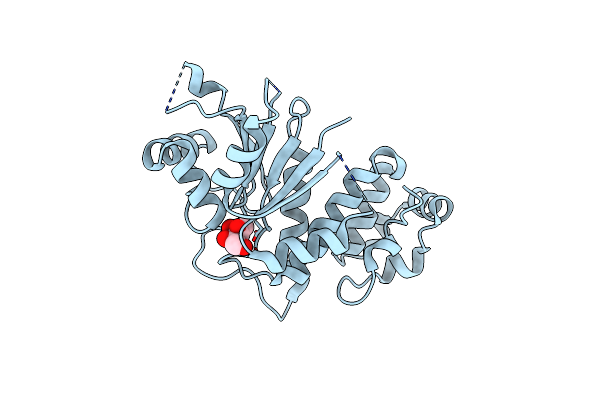 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-08-17 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: XYL |
 |
Crystal Structure Of Yeast Ynq8, A Fumarylacetoacetate Hydrolase Family Protein
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-06-15 Classification: Structural Genomics, Unknown Function Ligands: CA, SO4, ACY |
 |
Crystal Structure Of Saccharomyces Cerevisiae Nit3, A Member Of Branch 10 Of The Nitrilase Superfamily
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2001-10-04 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2001-09-26 Classification: structural genomics, unknown function Ligands: CL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2001-07-04 Classification: TRANSLATION INHIBITOR |
 |
Structural Studies Of Cholesterol Biosynthesis: Mevalonate 5-Diphosphate Decarboxylase And Isopentenyl Diphosphate Isomerase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-03-28 Classification: ISOMERASE Ligands: MN |
 |
The X-Ray Crystal Structure Of Mevalonate 5-Diphosphate Decarboxylase At 2.3 Angstrom Resolution.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2001-03-21 Classification: LYASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-09-02 Classification: STRUCTURAL GENOMICS Ligands: PLP |
 |
Crystal Structure Of A Yeast Hypothetical Protein-A Structure From Bnl'S Human Proteome Project
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1999-01-27 Classification: HYPOTHETICAL PROTEIN Ligands: PLP |
 |
The Structure Of Bacteriophage T7 Lysozyme, A Zinc Amidase And An Inhibitor Of T7 Rna Polymerase
Organism: Enterobacteria phage t7
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-04-30 Classification: HYDROLASE(ACTING ON LINEAR AMIDES) Ligands: ZN |

