Search Count: 7
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2021-04-28 Classification: DE NOVO PROTEIN Ligands: ZN, PEG, DMX, BEZ |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-04-21 Classification: DE NOVO PROTEIN Ligands: BEZ, 1PE, ZN, PEG, ACY |
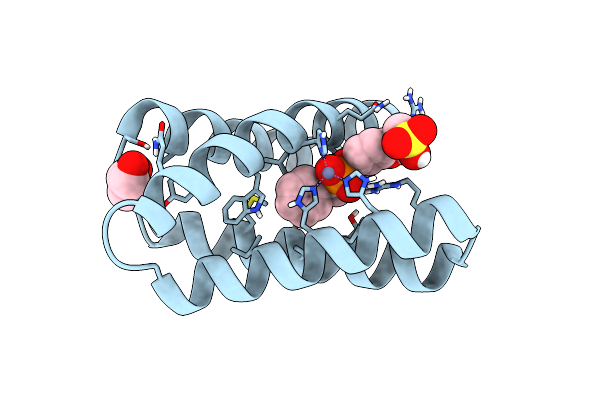 |
Structure Of The Engineered Metalloesterase Mid1Sc10 Complexed With A Phosphonate Transition State Analogue
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2018-12-12 Classification: De Novo Protein/Hydrolase Ligands: ZN, 9RQ, GOL |
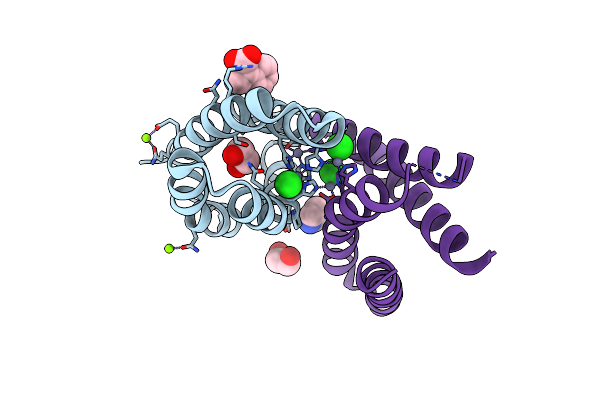 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2018-12-12 Classification: DE NOVO PROTEIN HYDROLASE Ligands: ZN, CL, MG, EDO, IMD, 9RW |
 |
Crystal Structure Of Retro-Aldolase Ra110.4-6 Complexed With Inhibitor 1-(6-Methoxy-2-Naphthalenyl)-1,3-Butanedione
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-11-12 Classification: ISOMERASE Ligands: LLK |
 |
Crystal Structure Of The Guanine Nucleotide Exchange Factor Sec12 (P1 Form)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2012-11-07 Classification: PROTEIN TRANSPORT Ligands: K |
 |
Crystal Structure Of The Guanine Nucleotide Exchange Factor Sec12 (P64 Form)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-11-07 Classification: PROTEIN TRANSPORT Ligands: K |

