Search Count: 84
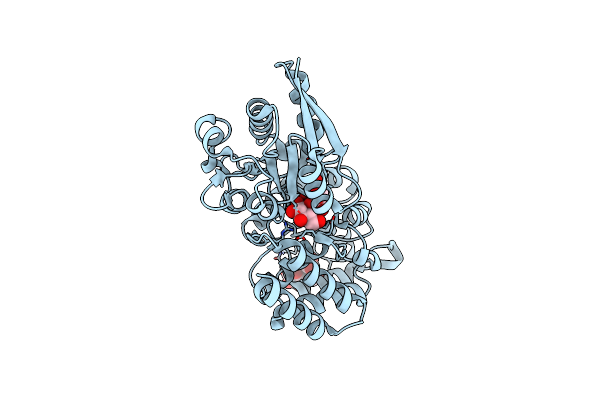 |
Crystal Structure Of Atu4361 Sugar Transporter From Agrobacterium Fabrum C58, Target Efi-510558, With Bound Sucrose
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2014-08-06 Classification: TRANSPORT PROTEIN |
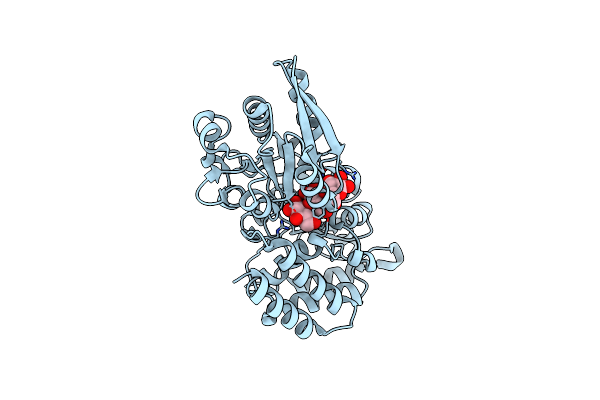 |
Crystal Structure Of Sugar Transporter Atu4361 From Agrobacterium Fabrum C58, Target Efi-510558, With Bound Maltotriose
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2014-07-16 Classification: TRANSPORT PROTEIN |
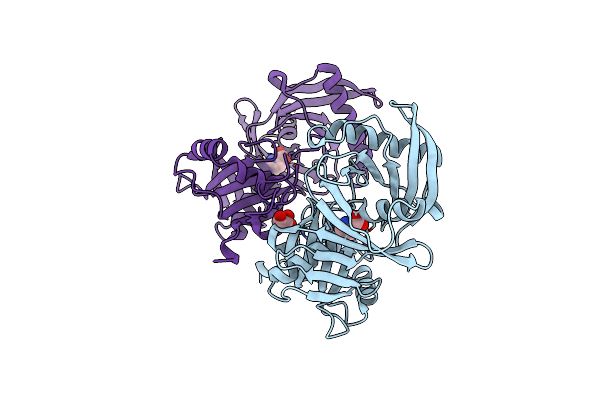 |
Crystal Structure Of A 4-Hydroxyproline Epimerase From Burkholderia Multivorans Atcc 17616, Target Efi-506586, Open Form, With Bound Pyrrole-2-Carboxylate
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-07 Classification: ISOMERASE Ligands: GOL, PYC |
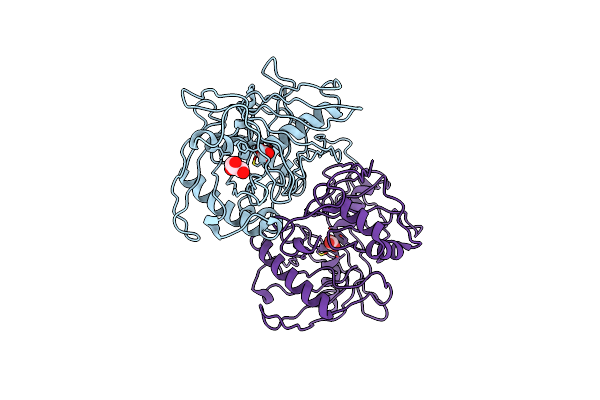 |
Crystal Structure Of Probable Proline Racemase From Agrobacterium Radiobacter K84, Target Efi-506561, With Bound Carbonate
Organism: Agrobacterium radiobacter k84
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-30 Classification: ISOMERASE Ligands: BCT, GOL |
 |
Crystal Structure Of Xylose Isomerase Domain Protein From Planctomyces Limnophilus Dsm 3776
Organism: Planctomyces limnophilus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-02-12 Classification: ISOMERASE Ligands: EDO |
 |
Crystal Structure Of A Hydroxyproline Epimerase From Agrobacterium Vitis, Target Efi-506420, With Bound Trans-4-Oh-L-Proline
Organism: Agrobacterium vitis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-17 Classification: ISOMERASE Ligands: ACT, HYP |
 |
Crystal Structure Of A 3-Hydroxyproline Dehydratse From Agrobacterium Vitis, Target Efi-506470, With Bound Pyrrole 2-Carboxylate, Ordered Active Site
Organism: Agrobacterium vitis s4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-01 Classification: ISOMERASE Ligands: CIT, ACT, PYC |
 |
Crystal Structure Of A Putative Hydroxyproline Epimerase From Xanthomonas Campestris (Target Efi-506516) With Bound Phosphate And Unknown Ligand
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-04-17 Classification: ISOMERASE Ligands: PO4, CL, UNL |
 |
Crystal Structure Of The Complex Of A Hydroxyproline Epimerase (Target Efi-506499, Pseudomonas Fluorescens Pf-5) With The Inhibitor Pyrrole-2-Carboxylate
Organism: Pseudomonas protegens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-03-13 Classification: Isomerase/Isomerase Inhibitor complex Ligands: GOL, PYC, NA |
 |
Crystal Structure Of The Complex Of A Hydroxyproline Epimerase (Target Efi-506499, Pseudomonas Fluorescens Pf-5) With Trans-4-Hydroxy-L-Proline
Organism: Pseudomonas protegens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-03-13 Classification: ISOMERASE Ligands: HYP, NA |
 |
Crystal Structure Of Pput_1285, A Putative Hydroxyproline Epimerase From Pseudomonas Putida F1 (Target Efi-506500), Open Form, Space Group I2, Bound Citrate
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2013-03-13 Classification: ISOMERASE Ligands: CIT |
 |
Crystal Structure Of Csal_2705, A Putative Hydroxyproline Epimerase From Chromohalobacter Salexigens (Target Efi-506486), Space Group P212121, Unliganded
Organism: Chromohalobacter salexigens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-03-13 Classification: ISOMERASE |
 |
Crystal Structure Of Pput_1285, A Putative Hydroxyproline Epimerase From Pseudomonas Putida F1 (Target Efi-506500), Open Form, Space Group P212121, Bound Sulfate
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-03-06 Classification: ISOMERASE Ligands: SO4 |
 |
Crystal Structure Of A Proposed Galactarolactone Cycloisomerase From Agrobacterium Tumefaciens, Target Efi-500704, With Bound Ca, Ordered Loops
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-11-07 Classification: ISOMERASE Ligands: CA, IMD, NI |
 |
Crystal Structure Of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis Htcc2601, With Bound Mg And L-4-Hydroxyproline Betaine (Betonicine)
Organism: Pelagibaca bermudensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-10 Classification: ISOMERASE Ligands: MG, NI, 0XW, IOD, MPD |
 |
Crystal Structure Of Efi-502318, An Enolase Family Member From Agrobacterium Tumefaciens With Homology To Dipeptide Epimerases (Bound Sodium, L-Ala-L-Glu With Ordered Loop)
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-08-29 Classification: ISOMERASE |
 |
Crystal Structure Of A Proposed Galactarolactone Cycloisomerase From Agrobacterium Tumefaciens, Target Efi-500704, With Bound Ca, Disordered Loops
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-08-29 Classification: ISOMERASE Ligands: CA, CL |
 |
Crystal Structure Of Beta-Phosphoglucomutase Homolog From Escherichia Coli, Target Efi-501172, With Bound Mg, Open Lid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-08-15 Classification: ISOMERASE Ligands: MG, CL, SO4, UNL, GOL |
 |
Crystal Structure Of Mannonate Dehydratase Prk15072 (Target Efi-502214) From Caulobacter Sp. K31
Organism: Caulobacter sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-27 Classification: ISOMERASE Ligands: MG, GOL, CL, UNL |
 |
Crystal Structure Of An Enolase (Mandelate Racemase Subgroup Member) From Agrobacterium Tumefaciens (Target Efi-502088) With Bound Mg And Formate
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2012-03-07 Classification: ISOMERASE Ligands: MG, FMT, CL |

