Search Count: 31
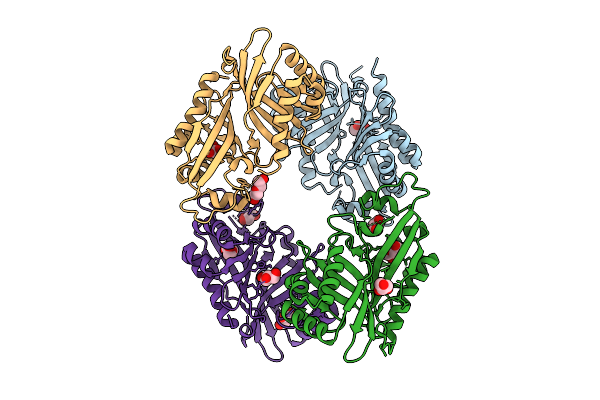 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL, PEG |
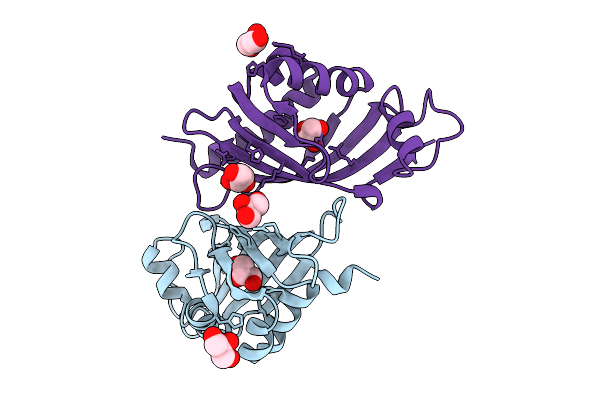 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL |
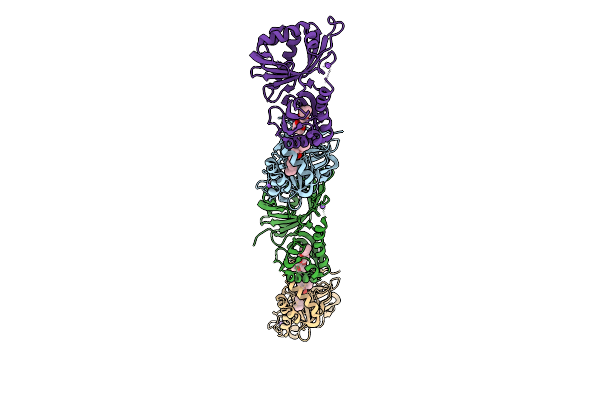 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1L6P, NA |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-16 Classification: FLAVOPROTEIN Ligands: FAD, Y7R, CL |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-04-16 Classification: FLAVOPROTEIN Ligands: FAD, GOL, YGK, CL |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-04-16 Classification: FLAVOPROTEIN Ligands: FAD, YK6, GOL, CL |
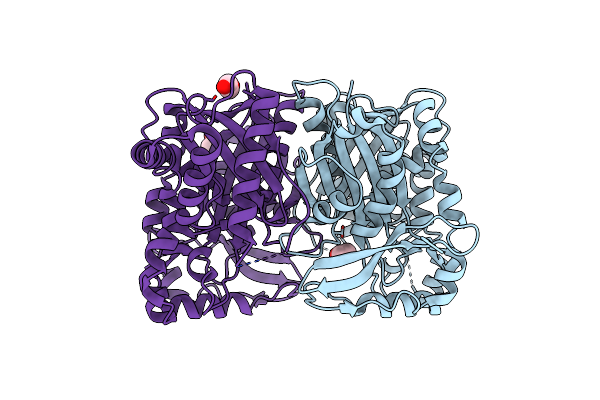 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: EDO |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Organism: Escherichia coli, Streptomyces virginiae
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: RIBOSOME Ligands: MUL |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-15 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, CL, NA, PNS |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-03-15 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, IMD, CL |
 |
Organism: Escherichia coli, Streptomyces virginiae
Method: ELECTRON MICROSCOPY Release Date: 2020-06-17 Classification: RIBOSOME Ligands: O7V |
 |
Organism: Escherichia coli, Streptomyces virginiae
Method: ELECTRON MICROSCOPY Release Date: 2020-06-17 Classification: RIBOSOME Ligands: O7S |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2016-03-23 Classification: PROTEIN BINDING |
 |
1H, 13C, 15N Chemical Shift Assignments Of Streptomyces Virginiae Vira Acp5A
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2014-06-04 Classification: TRANSFERASE |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2014-06-04 Classification: RIBOSOMAL PROTEIN |
 |
Crystal Structure Of Virginiamycin M And S Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui, Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-04-26 Classification: RIBOSOME/ANTIBIOTIC Ligands: MG, K, NA, CL, VIR, CD |

