Search Count: 22
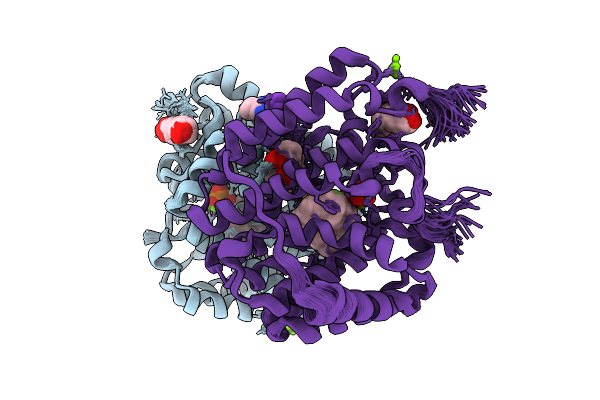 |
Ensemble Refined Structure Of Cotb2 In Complex With 2-Fluoro-3,7,18-Dolabellatriene
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: HYDROLASE Ligands: MG, PPV, EXW, MPD, CL, CCN, 2PE, K |
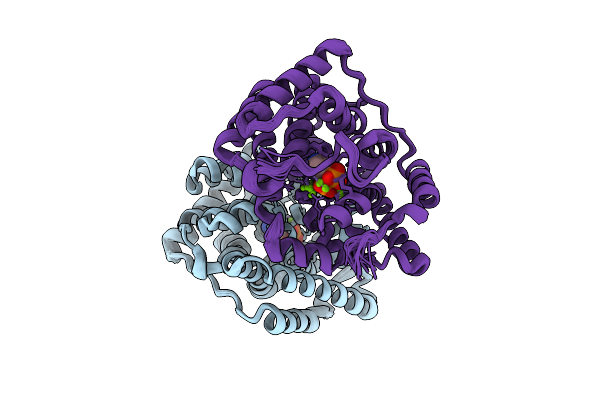 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: HYDROLASE Ligands: AHD, MG |
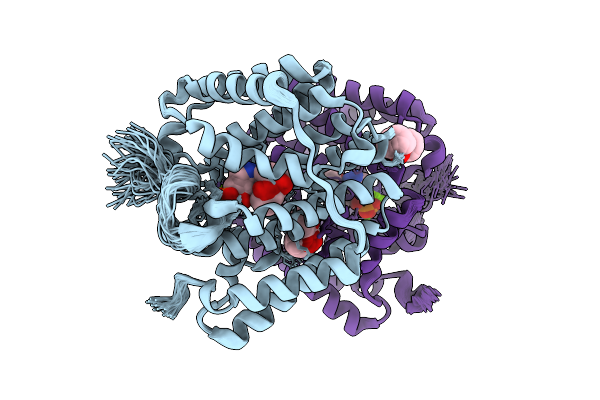 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: HYDROLASE Ligands: MPD, AHD, MG, CL |
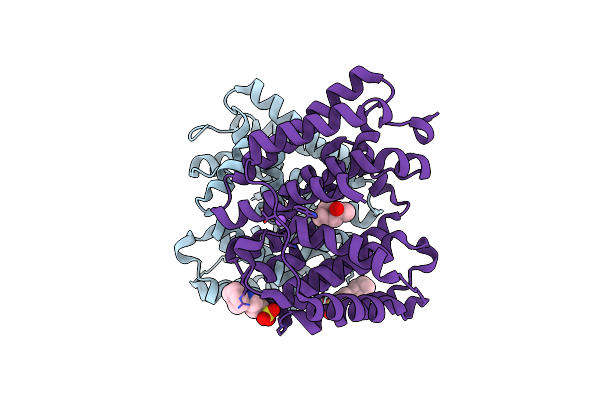 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-24 Classification: LYASE Ligands: CXS, NA, MPD |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-04-24 Classification: LYASE Ligands: AHD, MG, EDO, MPD, ACT, PG0, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-12-02 Classification: LYASE Ligands: AHD, MPD, MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: LYASE Ligands: AHD, MG, CL, MPD |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-12-02 Classification: LYASE Ligands: MPD, AHD, MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-12-02 Classification: LYASE Ligands: AHD, MPD, MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-12-02 Classification: LYASE Ligands: MPD |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-12-02 Classification: LYASE Ligands: MPD, MG |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-10 Classification: LYASE Ligands: MG, PPV, EXW, MPD, CL, CCN, PEG, K |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-10-10 Classification: LYASE Ligands: AHD, MG |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-10-10 Classification: LYASE Ligands: MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-10-10 Classification: LYASE Ligands: MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-06-07 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT |
 |
Crystal Structure Of Cotb2 (Ggspp/Mg2+-Bound Form) From Streptomyces Melanosporofaciens
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-06-07 Classification: BIOSYNTHETIC PROTEIN Ligands: GGS, MG |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2014-06-04 Classification: LYASE Ligands: CXS, NA |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2014-06-04 Classification: LYASE Ligands: CXS, NA |
 |
NA
Organism: Streptomyces melanosporofaciens
Method: Alphafold Release Date: Classification: NA Ligands: NA |

