Search Count: 25
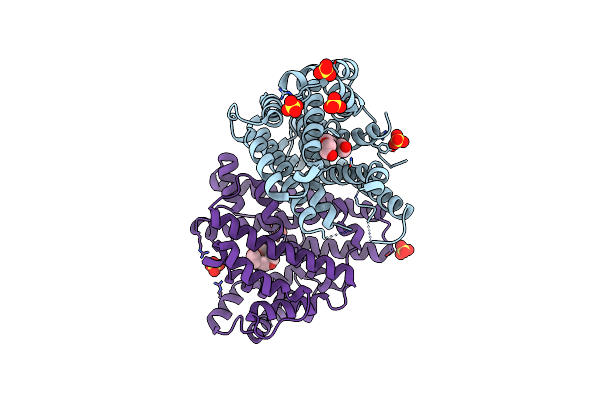 |
Crystal Structure Of Pentalenene Synthase Variant F76A With Peg Molecule In The Active Site
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: PG4, SO4 |
 |
Crystal Structure Of Pentalenene Synthase Variant F76A Complexed With 2-Fluorofarnesyl Diphosphate
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: FPF, MG |
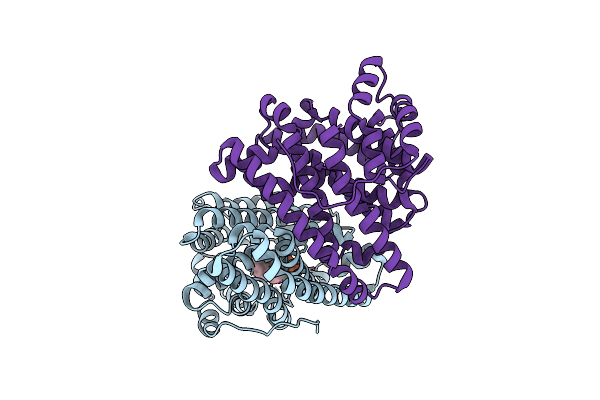 |
Crystal Structure Of Pentalenene Synthase Variant F76A Complexed With 12,13-Difluorofarnesyl Diphosphate
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: FDF, MG |
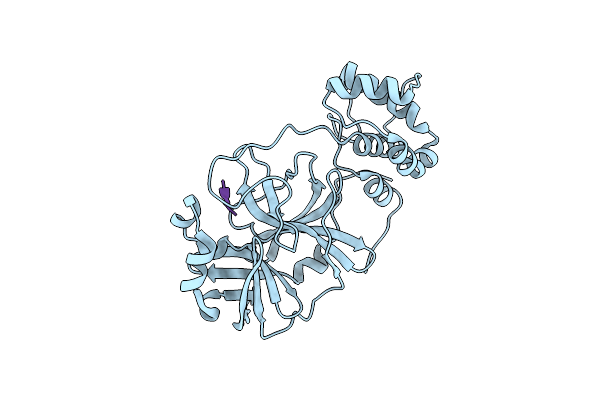 |
Room-Temperature X-Ray Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Leupeptin
Organism: Severe acute respiratory syndrome coronavirus 2, Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-06-17 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR |
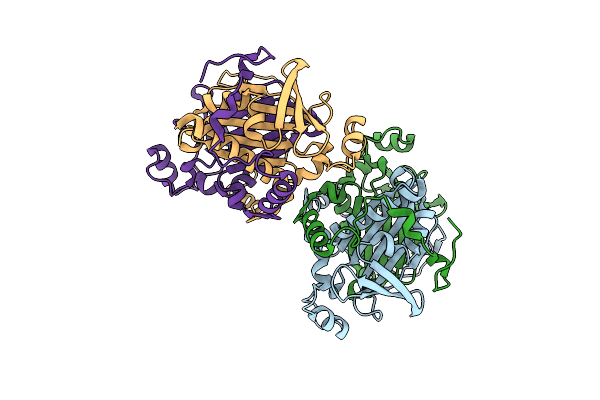 |
Crystal Structure Of Cqsb2 From Streptomyces Exfoliatus 2419-Svt2 (Apo Form)
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Cqsb2 From Streptomyces Exfoliatus In Complex With The Product, 1-(2-Hydroxypropyl)-2-Methyl-Carbazole-3,4-Dione
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-16 Classification: BIOSYNTHETIC PROTEIN Ligands: AO6 |
 |
Structural, Thermodynamic And Kinetic Analysis Of The Picomolar Binding Affinity Interaction Of The Beta-Lactamase Inhibitor Protein-Ii (Blip-Ii) With Class A Beta-Lactamases
Organism: Bacillus anthracis, Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2011-07-20 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Structural, Thermodynamic And Kinetic Analysis Of The Picomolar Binding Affinity Interaction Of The Beta-Lactamase Inhibitor Protein-Ii (Blip-Ii) With Class A Beta-Lactamases
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-07-20 Classification: HYDROLASE INHIBITOR Ligands: SO4 |
 |
Crystal Structure Of Beta-Lactamse Inhibitory Protein-I (Blip-I) In Apo Form
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-03-31 Classification: PROTEIN BINDING Ligands: TAM |
 |
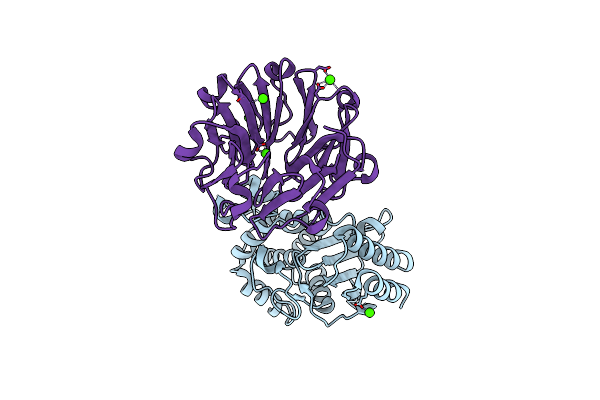
Crystal Structure Of Beta-Lactamse Inhibitory Protein-I (Blip-I) In Complex With Tem-1 Beta-Lactamase
Organism: Escherichia sp. sflu5, Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-03-31 Classification: PROTEIN BINDING Ligands: PO4 |
 |
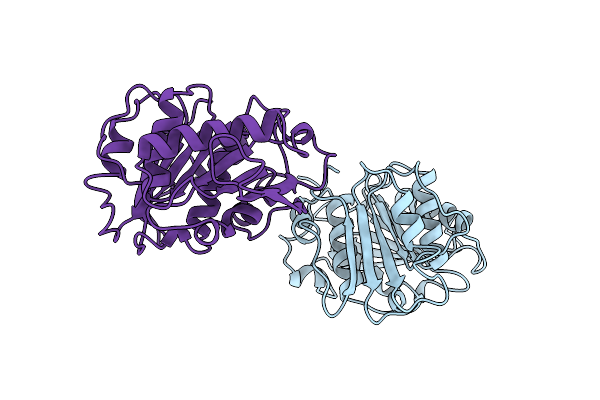
Crystal Structure Of Beta-Lactamase Inhibitor Protein-Ii In Complex With Tem-1 Beta-Lactamase
Organism: Escherichia coli, Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2001-10-03 Classification: HYDROLASE/INHIBITOR Ligands: CA |
 |
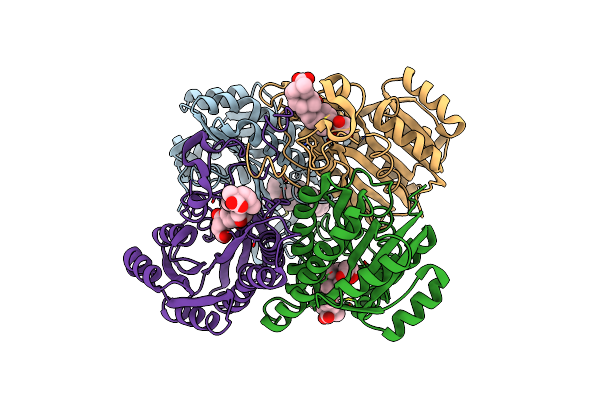
Crystal Structure Of The Streptomyces Exfoliatus Lipase At 1.9A Resolution: A Model For A Family Of Platelet-Activating Factor Acetylhydrolases
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1998-07-15 Classification: SERINE HYDROLASE |
 |
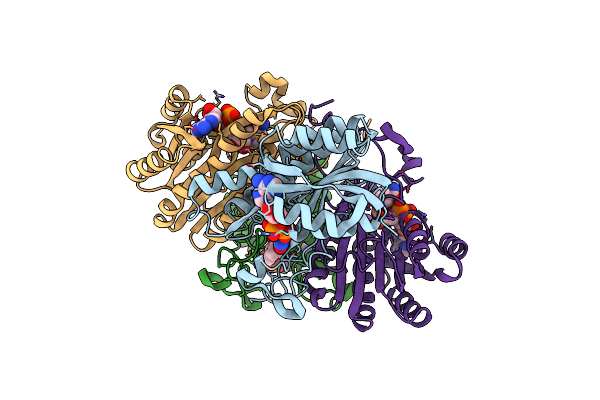
Mechanism Of Inhibition Of 3Alpha,20Beta-Hydroxysteroid Dehydrogenase By A Licorice-Derived Steroidal Inhibitor
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1995-02-07 Classification: OXIDOREDUCTASE Ligands: CBO |
 |
The Refined Three-Dimensional Structure Of 3Alpha,20Beta-Hydroxysteroid Dehydrogenase And Possible Roles Of The Residues Conserved In Short-Chain Dehydrogenases
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 1994-08-31 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
NA
|
 |
NA
|
 |
NA
|
 |
NA
|
 |
NA
|
 |
NA
|

