Search Count: 50
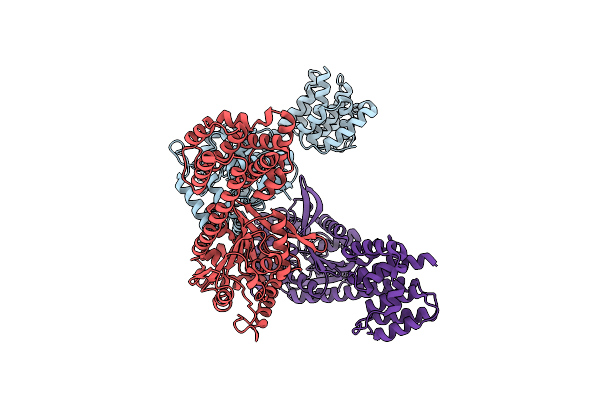 |
Structure Of A 16 Nm Protein Cage Designed By Fusing Symmetric Oligomeric Domains, Quadruple Mutant, I222 Form
Organism: Streptomyces aureofaciens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:4.19 Å Release Date: 2015-05-20 Classification: OXIDOREDUCTASE, VIRAL PROTEIN |
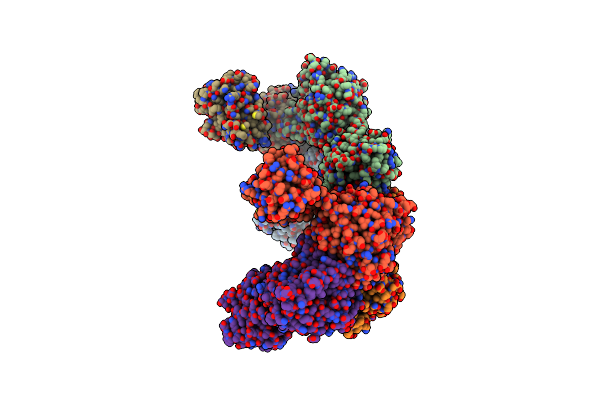 |
Structure Of A 16 Nm Protein Cage Designed By Fusing Symmetric Oligomeric Domains, Quadruple Mutant, P21212 Form
Organism: Streptomyces aureofaciens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:6.49 Å Release Date: 2015-05-20 Classification: OXIDOREDUCTASE, VIRAL PROTEIN |
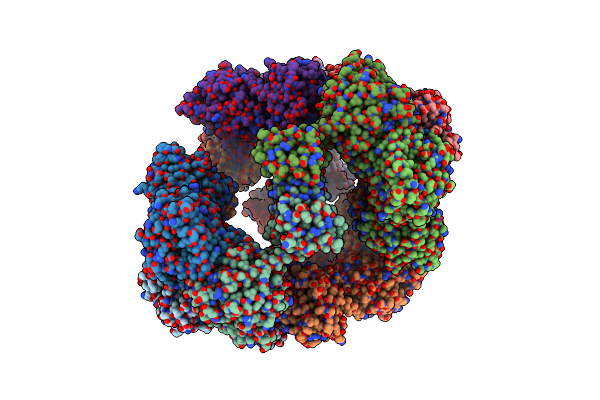 |
Structure Of A 16 Nm Protein Cage Designed By Fusing Symmetric Oligomeric Domains, Quadruple Mutant, P212121 Form
Organism: Streptomyces aureofaciens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:7.81 Å Release Date: 2015-05-20 Classification: OXIDOREDUCTASE, VIRAL PROTEIN |
 |
Crystal Structure Analysis Of Streptomyces Aureofaciens Ribonuclease Sa T95A Mutant
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2014-05-28 Classification: HYDROLASE Ligands: SO4, GOL, CAC |
 |
Crystal Structure Analysis Of Streptomyces Aureofaciens Ribonuclease Sa Y51F Mutant
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2014-05-28 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Analysis Of Streptomyces Aureofaciens Ribonuclease S24A Mutant
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2013-08-14 Classification: HYDROLASE Ligands: SO4 |
 |
Structure Of A 16 Nm Protein Cage Designed By Fusing Symmetric Oligomeric Domains, Triple Mutant, P21212 Form
Organism: Streptomyces aureofaciens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2013-07-24 Classification: OXIDOREDUCTASE |
 |
Structure Of A 16 Nm Protein Cage Designed By Fusing Symmetric Oligomeric Domains, Triple Mutant, P212121 Form
Organism: Streptomyces aureofaciens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2013-07-24 Classification: OXIDOREDUCTASE |
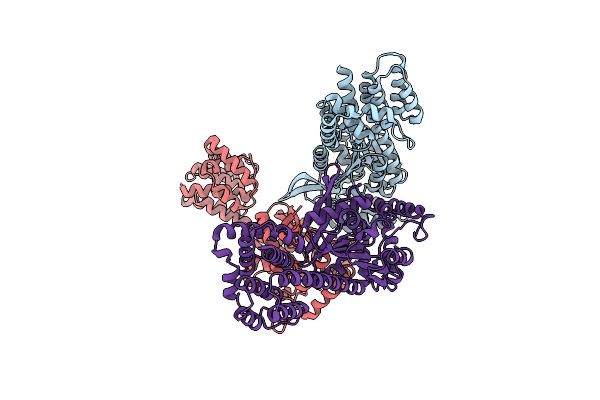 |
Structure Of A 16 Nm Protein Cage Designed By Fusing Symmetric Oligomeric Domains, Triple Mutant, I222 Form
Organism: Streptomyces aureofaciens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:7.35 Å Release Date: 2013-07-24 Classification: OXIDOREDUCTASE |
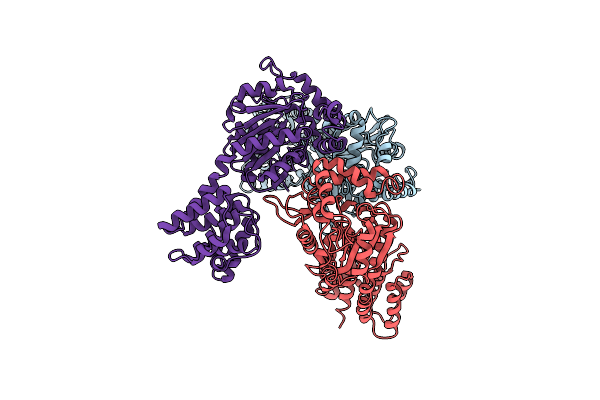 |
Structure Of A 16 Nm Protein Cage Designed By Fusing Symmetric Oligomeric Domains
Organism: Streptomyces aureofaciens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-06-20 Classification: DE NOVO PROTEIN |
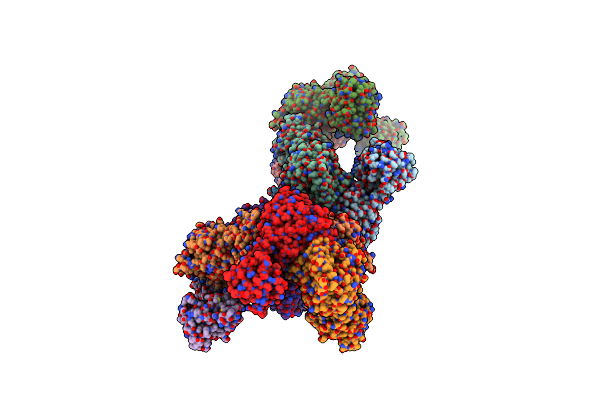 |
Structure Of A 16 Nm Protein Cage Designed By Fusing Symmetric Oligomeric Domains
Organism: Streptomyces aureofaciens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:3.92 Å Release Date: 2012-06-20 Classification: DE NOVO PROTEIN |
 |
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-08-04 Classification: HYDROLASE |
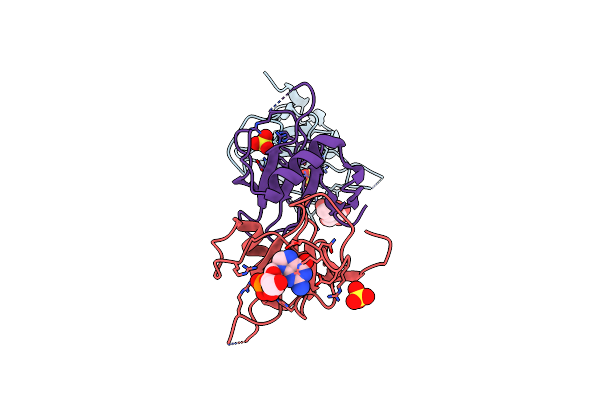 |
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-06-16 Classification: HYDROLASE Ligands: BGC, SO4, 2GP |
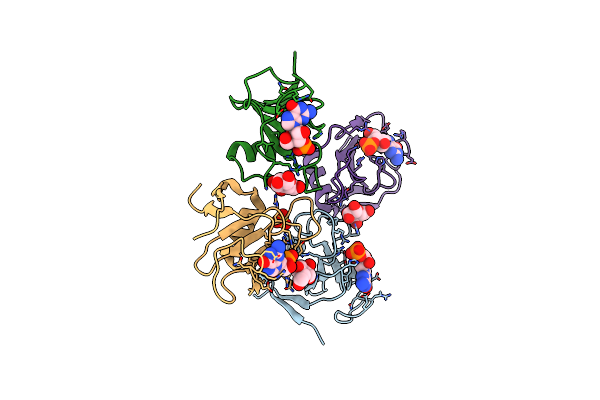 |
Crystal Structure Of Ribonuclease Sa2 With Guanosine-3'-Cyclophosphate Prepared By Cocrystallization
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-06-16 Classification: HYDROLASE Ligands: BGC, SO4, 3GP |
 |
Crystal Structure Of Ribonuclease Sa2 With 3'-Gmp Obtained By Ligand Diffusion
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-05-26 Classification: HYDROLASE Ligands: SO4, 3GP |
 |
Structure Of Ribonuclease Sa2 Complexes With Mononucleotides: New Aspects Of Catalytic Reaction And Substrate Recognition
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-05-26 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-05-26 Classification: HYDROLASE Ligands: SO4, SGP |
 |
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2006-08-08 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2005-07-19 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Streptomyces aureofaciens
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2004-12-21 Classification: HYDROLASE Ligands: SO4 |

