Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
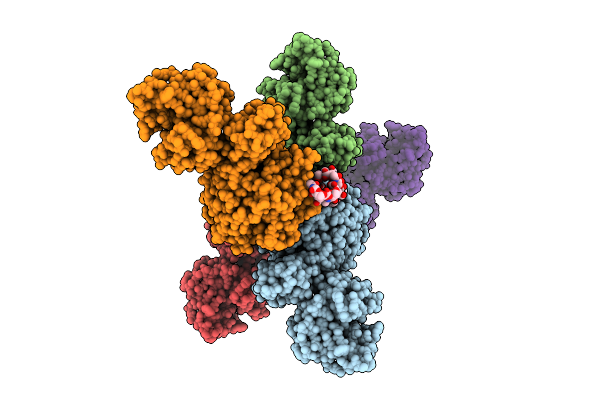 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TOXIN Ligands: A2G, A1CAY, A1CAZ |
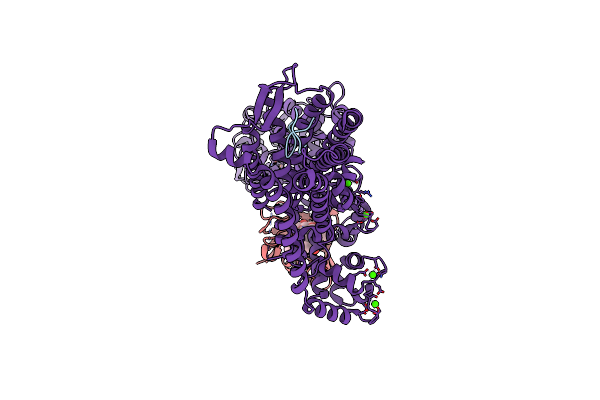 |
Cryoem Structure Of Calcineurin-Fusion Human Endothelin Receptor Type-B In Complex With Res-701-3
Organism: Homo sapiens, Streptomyces venezuelae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: MEMBRANE PROTEIN Ligands: CA, FE, ZN, FK5 |
 |
Streptomyces Griseus Family 2B Encapsulin Shell With 2-Methylisoborneol Synthase Cargo In 20 Mm Camp
Organism: Streptomyces griseus subsp. griseus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: VIRUS LIKE PARTICLE Ligands: CA |
 |
Streptomyces Griseus Family 2B Encapsulin Shell With 2-Methylisoborneol Synthase Cargo
Organism: Streptomyces griseus subsp. griseus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: VIRUS LIKE PARTICLE Ligands: CA |
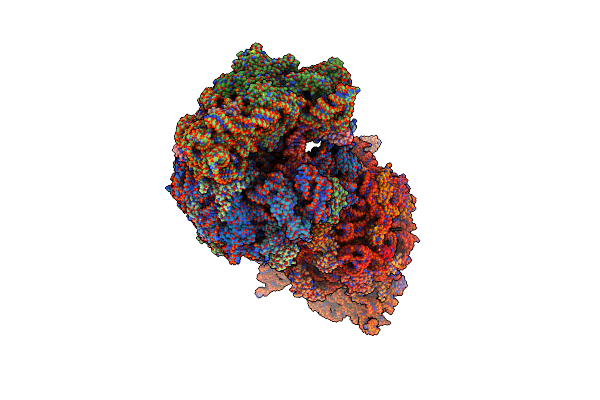 |
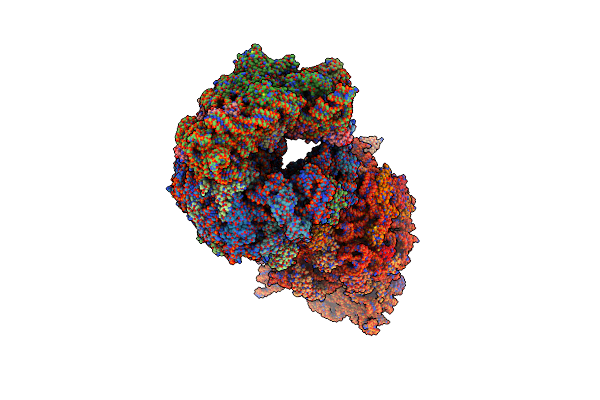
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
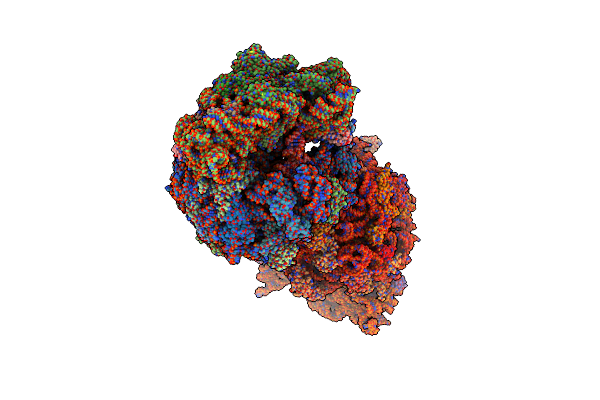 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin B, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.60A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin And Protein Y At 2.60A Resolution
Organism: Escherichia coli k-12, Paenibacillus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ARG, MPD, ZN, SF4 |
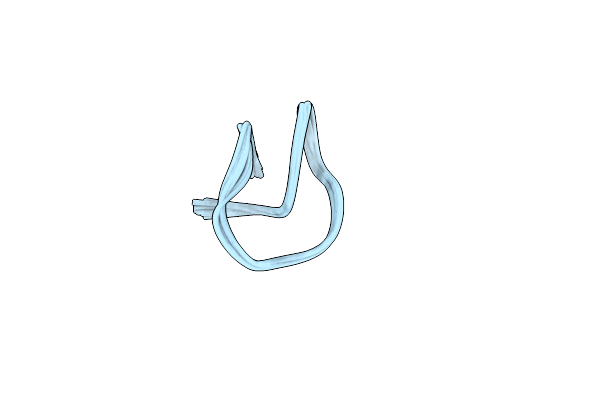 |
Organism: Lentzea jiangxiensis
Method: SOLUTION NMR Release Date: 2024-09-25 Classification: UNKNOWN FUNCTION |
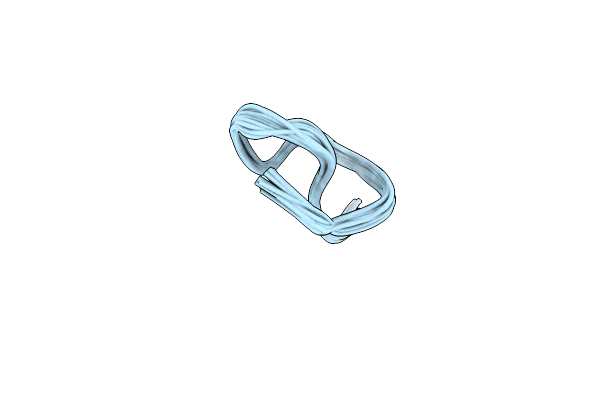 |
Organism: Streptomyces katrae
Method: SOLUTION NMR Release Date: 2024-09-25 Classification: UNKNOWN FUNCTION |
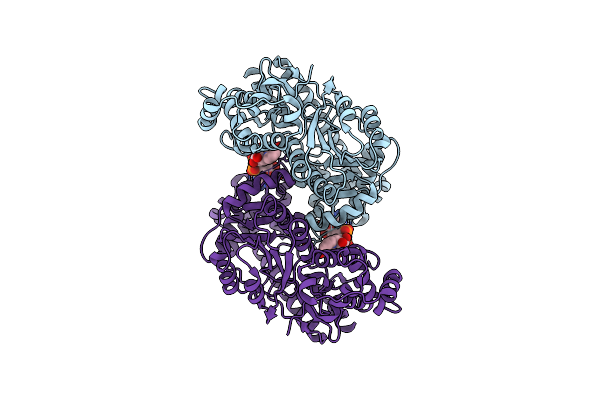 |
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: LYASE Ligands: NAD |
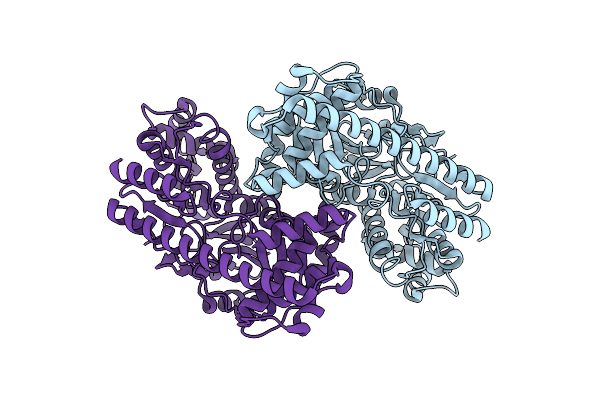 |
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: LYASE |
 |
Organism: Streptomyces noursei atcc 11455
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Streptosporangium roseum, Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: PEG |
 |
Organism: Streptosporangium roseum, Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: PEG, ATP |
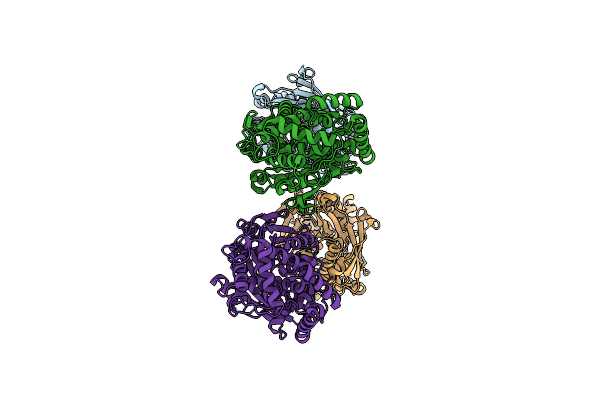 |
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN |
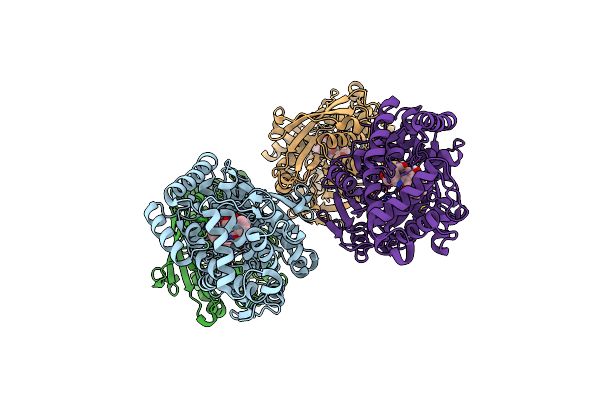 |
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN Ligands: 9VG |
 |
Crystal Structure Of Sznf From Streptomyces Achromogenes Var. Streptozoticus Nrrl 2697 Mononuclear Fe(Ii) Structure On The Hdo Cofactor Assembly Pathway
Organism: Streptomyces achromogenes subsp. streptozoticus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: FE2 |
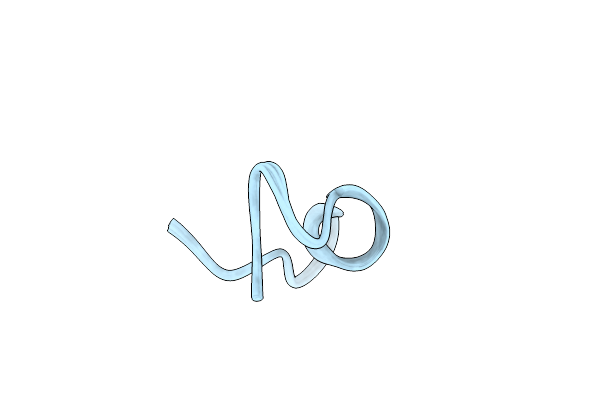 |
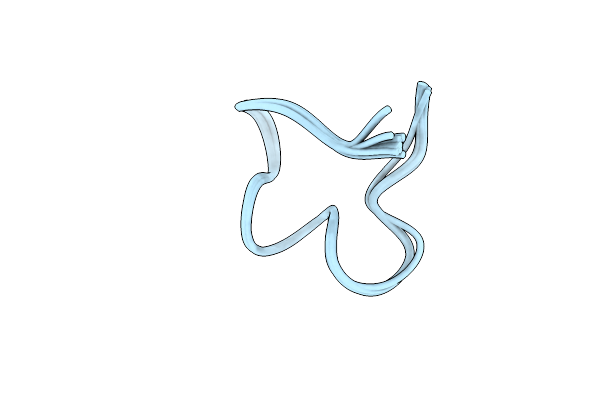
The Nmr Structure Of Noursin, A Tricyclic Ribosomal Peptide Containing A Histidine-To-Butyrine Crosslink
Organism: Streptomyces noursei atcc 11455
Method: SOLUTION NMR Release Date: 2023-05-31 Classification: UNKNOWN FUNCTION |
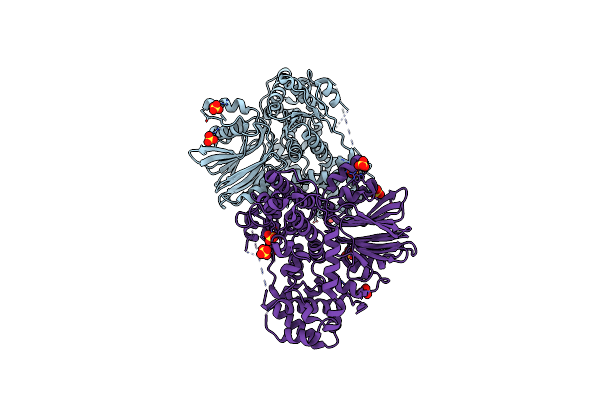 |
Organism: Streptomyces sp. rm72
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2023-05-31 Classification: LIGASE Ligands: SO4 |
 |
Organism: Streptomyces noursei atcc 11455
Method: SOLUTION NMR Release Date: 2023-05-31 Classification: UNKNOWN FUNCTION |