Search Count: 11
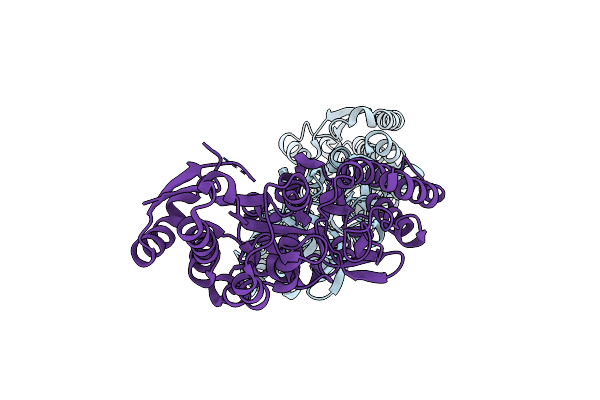 |
Crystal Structure Of Maltodextrin-Binding Protein Sps0871 From Streptococcus Pyogenes At 2.20 A
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: SUGAR BINDING PROTEIN |
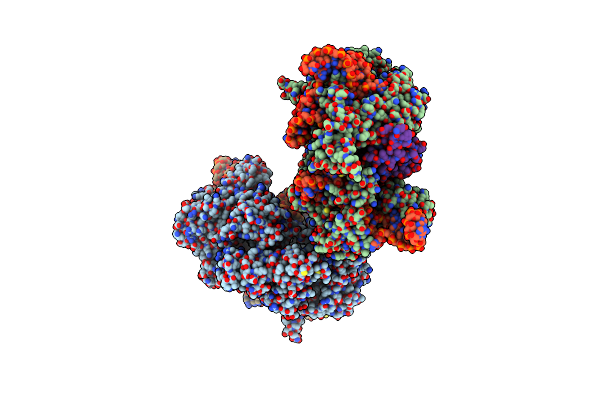 |
Organism: Streptococcus pyogenes serotype m1, Listeria monocytogenes, Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2019-01-23 Classification: RNA BINDING PROTEIN/RNA |
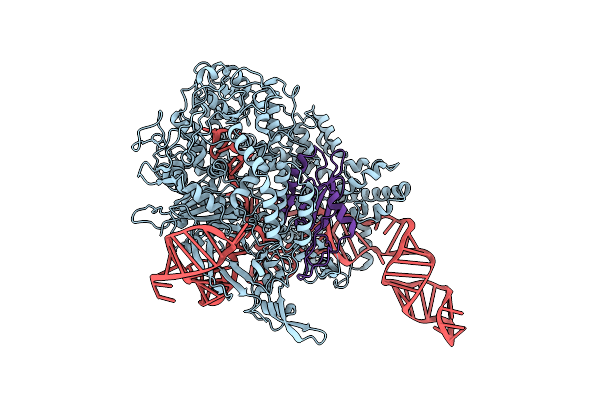 |
Organism: Streptococcus pyogenes m1 gas, Listeria phage lp-101
Method: ELECTRON MICROSCOPY Release Date: 2019-01-16 Classification: HYDROLASE/RNA/VIRAL PROTEIN |
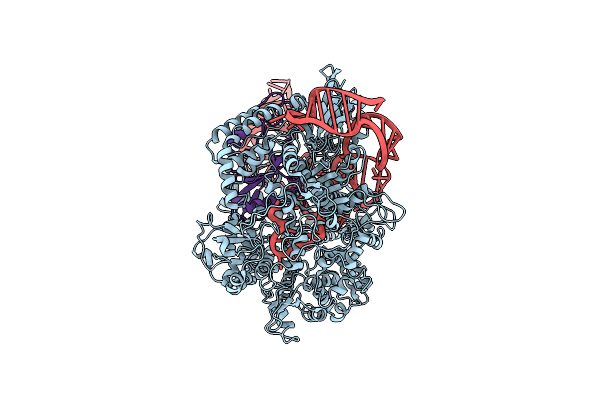 |
Organism: Streptococcus pyogenes m1 gas, Listeria phage lp-101
Method: ELECTRON MICROSCOPY Release Date: 2019-01-16 Classification: HYDROLASE/RNA/VIRAL PROTEIN |
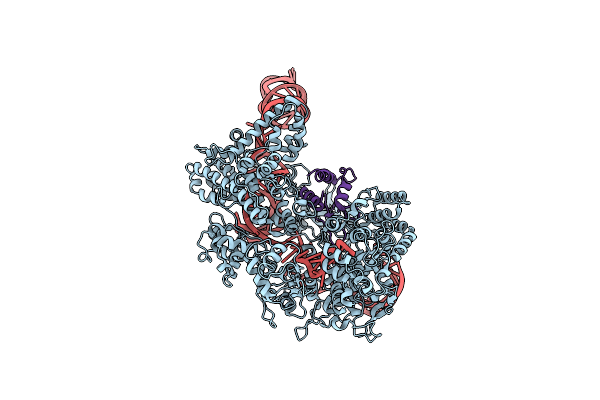 |
Organism: Streptococcus pyogenes m1 gas, Unidentified phage
Method: ELECTRON MICROSCOPY Release Date: 2017-07-26 Classification: Immune System/RNA |
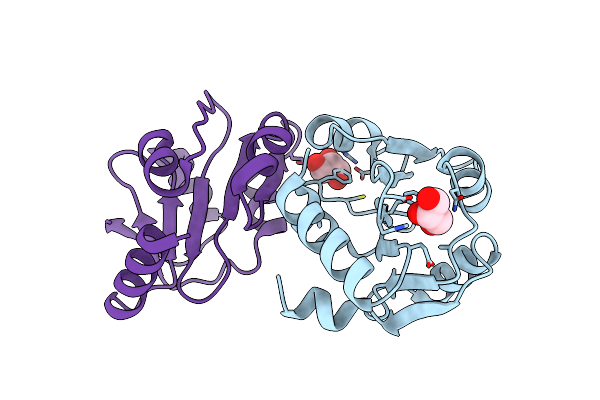 |
The Crystal Structure Of The Creatinase/Prolidase N-Terminal Domain Of An X-Pro Dipeptidase From Streptococcus Pyogenes To 1.85A
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-08-11 Classification: HYDROLASE Ligands: GOL, CL |
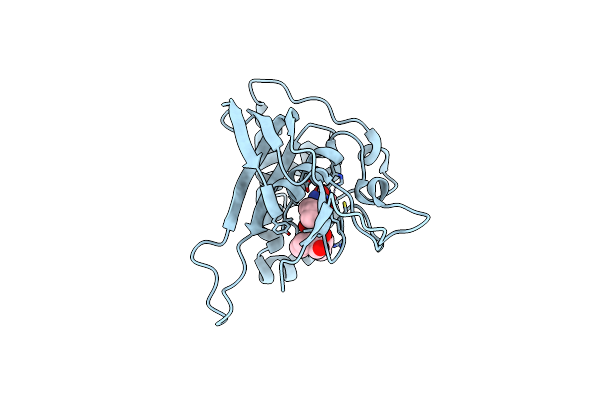 |
Structures Of Actinonin Bound Peptide Deformylases From E. Faecalis And S. Pyogenes
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2008-03-04 Classification: HYDROLASE Ligands: CO, BB2 |
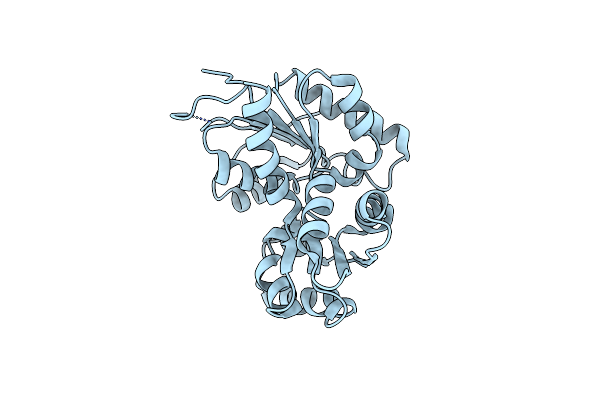 |
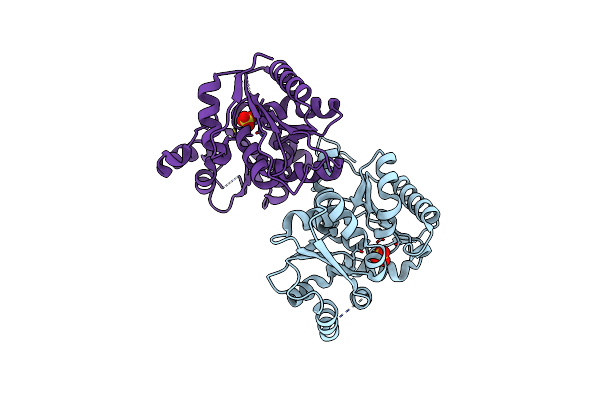
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-09-25 Classification: ISOMERASE |
 |
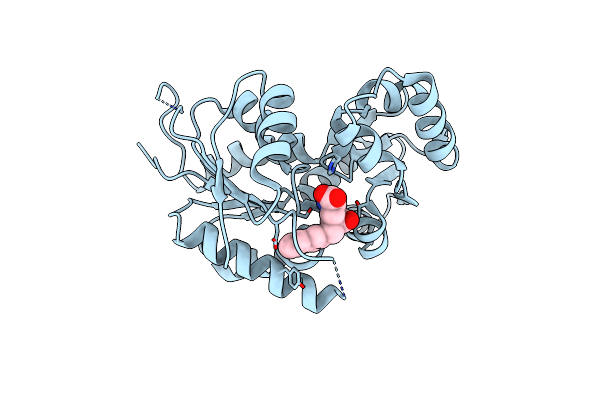
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2007-09-25 Classification: ISOMERASE Ligands: SO4 |
 |
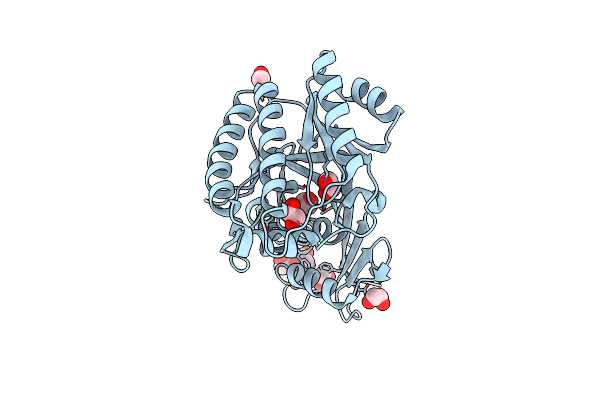
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-09-25 Classification: ISOMERASE Ligands: NHL |
 |
Structure Of Mevalonate Pyrophosphate Decarboxylase From Streptococcus Pyogenes
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-06-06 Classification: LYASE Ligands: EDO, ACY |

