Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
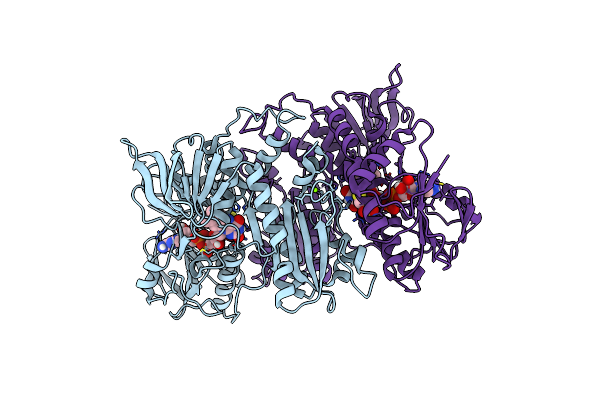 |
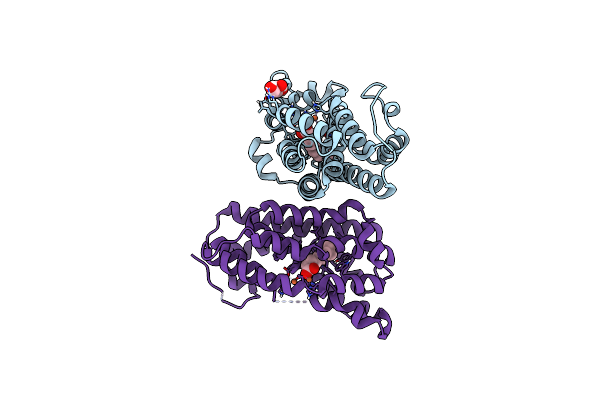
Crystal Structure Of F501H/H506E Variant Of 2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase (2-Kpcc) From Xanthobacter Autotrophicus
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: COM, FDA, MG |
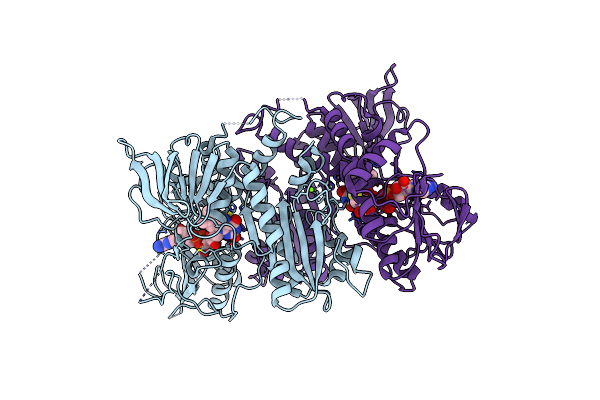 |
Crystal Structure Of F501H Variant Of 2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase (2-Kpcc) From Xanthobacter Autotrophicus
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: FDA, COM, MG |
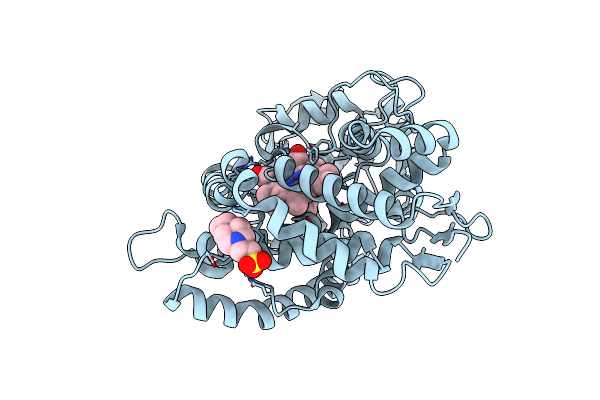 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEM, JZ3, EPE |
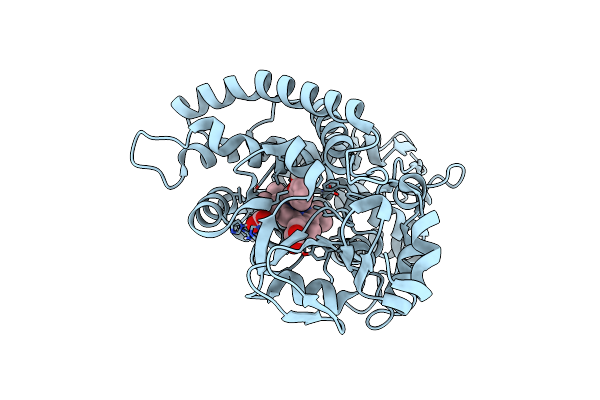 |
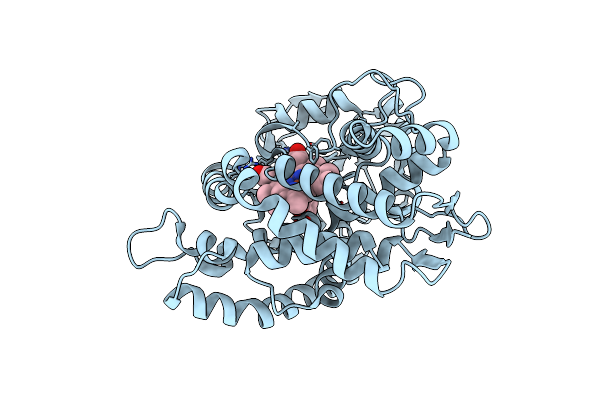
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEM, JZ3 |
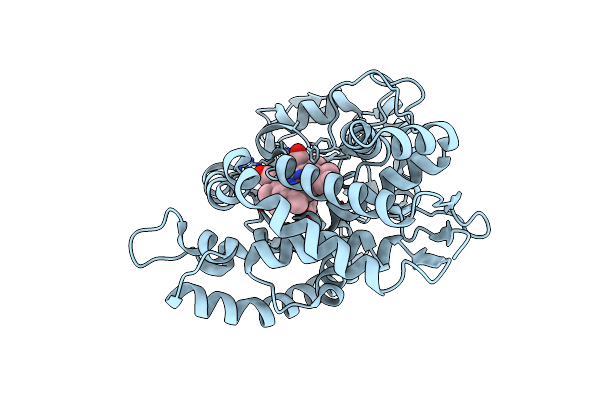 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEM, JZ3 |
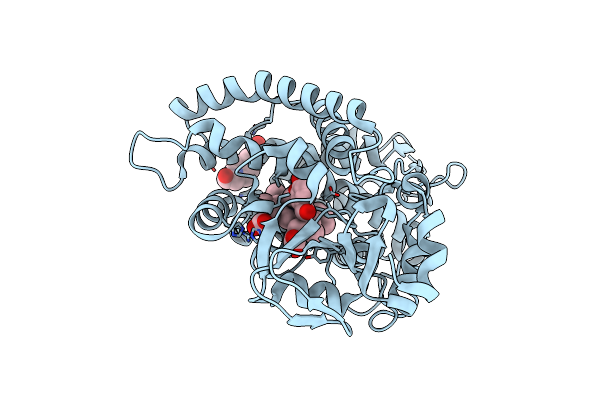 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEM, V55, EPE |
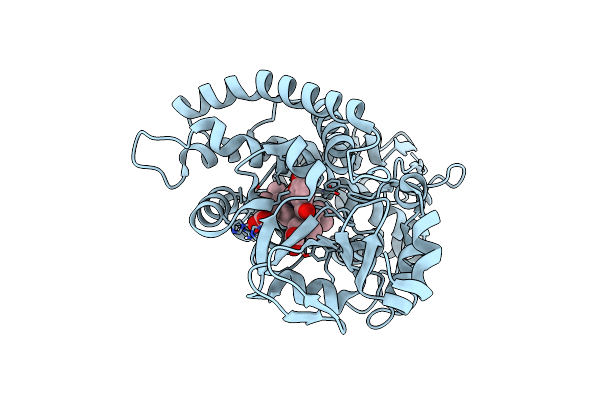 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: V55, HEM |
 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEM, V55 |
 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEM, OKE |
 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEM, OKE |
 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: HEM, OKE |
 |
Organism: Amycolatopsis sp. (strain atcc 39116 / 75iv2)
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: EPE, V55, HEC |
 |
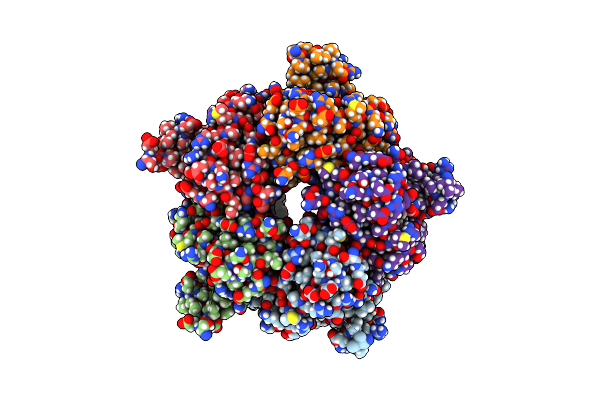
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: DAO, FE, GOL |
 |
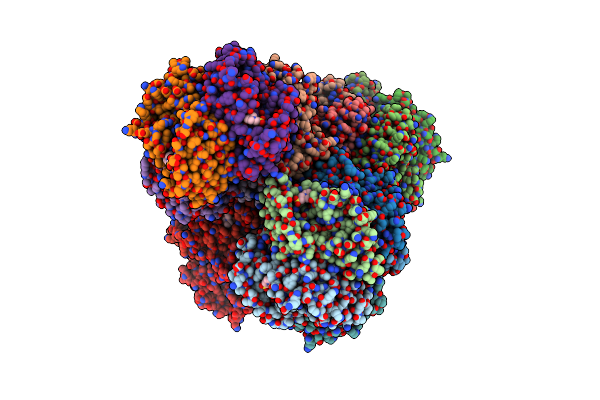
Organism: Geobacillus stearothermophilus 10
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-01-18 Classification: OXIDOREDUCTASE Ligands: 76R |
 |
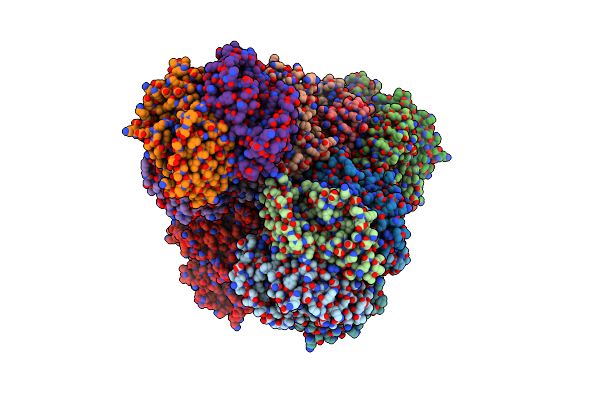
Organism: Dechloromonas aromatica
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2011-01-05 Classification: OXIDOREDUCTASE Ligands: HEM, NO2, MES, CA |
 |
Organism: Dechloromonas aromatica
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-01-05 Classification: OXIDOREDUCTASE Ligands: HEM, NO2, CA |