Search Count: 190
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: PROTEIN BINDING Ligands: A1I43, SO4 |
 |
Crystal Structure Of The Polyester Hydrolase Leipzig 7 (Phl7) Mut3 Variant With Glycosylation By Expression In Pichia Pastoris (P_Phl7Mut3)
Organism: Compost metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of The Non-Glycosylated Polyester Hydrolase Leipzig 7 (Phl7) Mut3 Variant Expressed In Pichia Pastoris (P_Phl7Mut3_Ng)
Organism: Compost metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of The Polyester Hydrolase Leipzig 7 (Phl7) Mut3 Variant Expressed In E. Coli (E_Phl7Mut3)
Organism: Compost metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHT, CL, MG |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17B
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHV, IMD |
 |
Crystal Structure Of Human Cd73 (Ecto-5'-Nucleotidase) In Complex With The Ampcp Analog Psb-12730 In The Closed State (Crystal Form Iv)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: ZN, A1IZZ |
 |
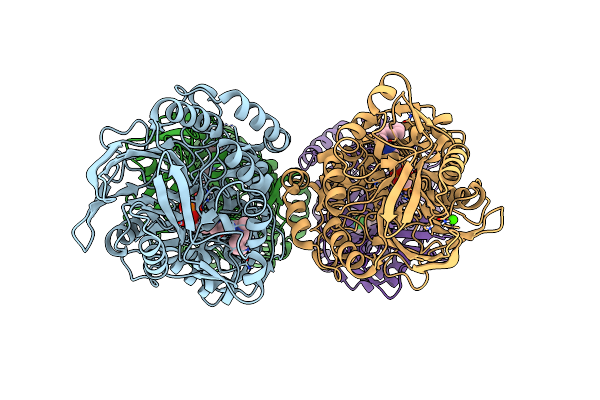
Crystal Structure Of Cd73 (Ecto-5'-Nucleotidase) Complexed To 8-Butylthioadenosine 5'Monophosphate (Compound 3 In Publication) In The Open Enzyme State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: ZN, CA, A1IVB, 1PE, PGE |
 |
Crystal Structure Of Human Cd73 (Ecto-5'-Nucleotidase) In Complex With An N6-Disubstituted Acyclic Adp Analog (Compound 26 In Publication) In The Closed State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: ZN, A1IVH, CL, CA |
 |
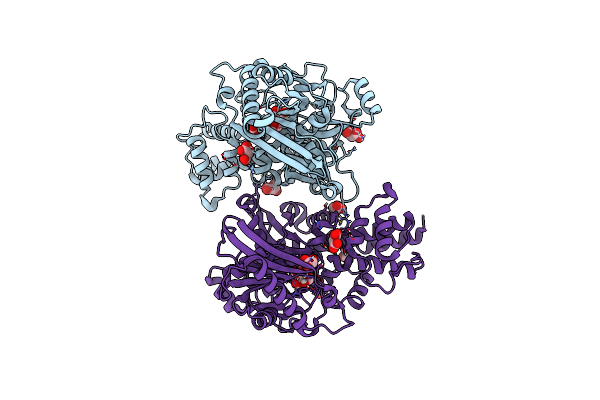
Crystal Structure Of Kluyveromyces Lactis Glucokinase In Complex With Glucose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: BGC, MLI |
 |
Crystal Structure Of Kluyveromyces Lactis Glucokinase In Complex With Mannose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MAN, MLI |
 |
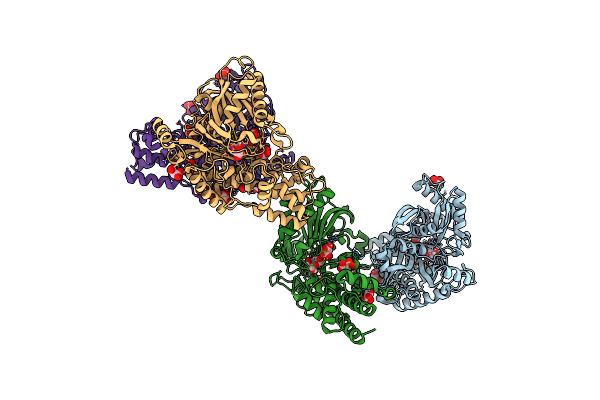
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Glucose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: BGC, MLI |
 |
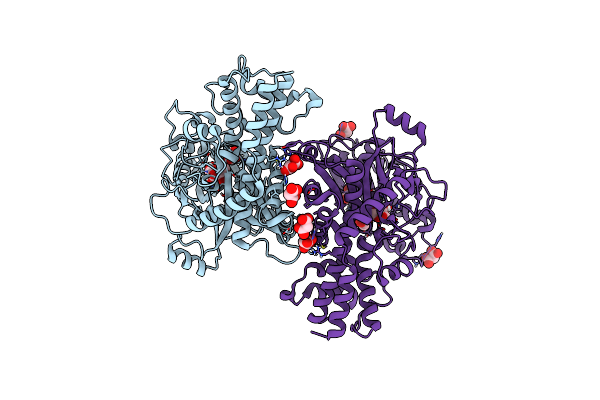
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Mannose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MAN, MLI |
 |
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Fructose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: FRU, MLI |
 |
Crystal Structure Of The Intermediate Open State Of Apo Kluyveromyces Lactis Glucokinase
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MLI |
 |
Crystal Structure Of The Open State Of Apo Kluyveromyces Lactis Glucokinase
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MLI |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Covalently Bound Inhibitor Fp237 (Compound 8P In Publication)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2025-01-29 Classification: VIRAL PROTEIN Ligands: A1IPK, DMS, PEG, EDO, NA, CL |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Covalently Bound Inhibitor Psb-21110 (Compound 29B In Publication)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-08-28 Classification: VIRAL PROTEIN Ligands: A1IF1, BR |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Covalently Bound Inhibitor Psb-21101 (Compound 30B In Publication)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-08-28 Classification: VIRAL PROTEIN Ligands: A1IEY, MG, BR |
 |
Human Ppargamma Ligand Binding Domain In Complex With Co-Activator 1Alpha Peptide And Bisphenol A (Bpa)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-07-10 Classification: TRANSCRIPTION Ligands: 2OH |

