Search Count: 53
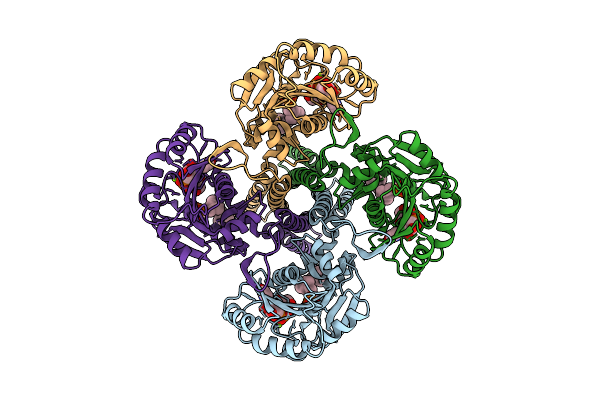 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
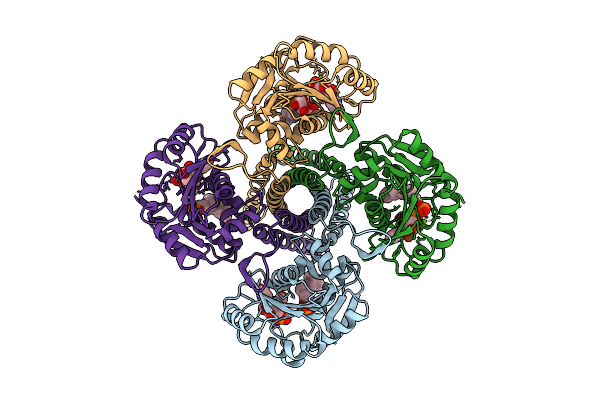 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
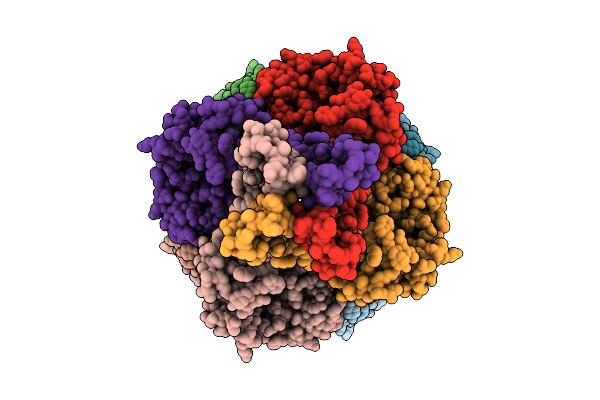 |
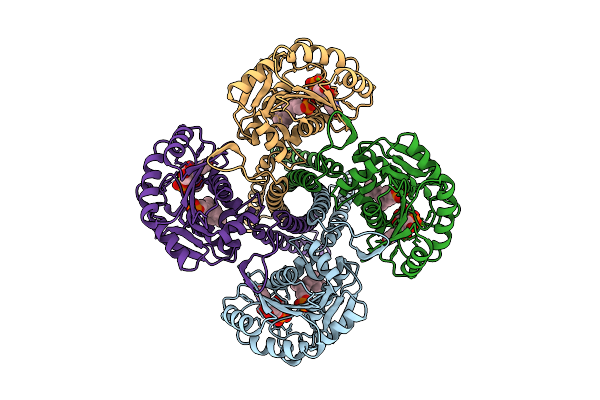
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
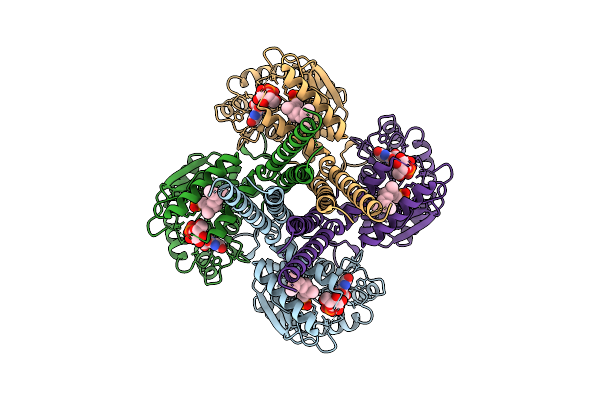 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN Ligands: 6WT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: PROTEIN FIBRIL |
 |
Intact V-Atpase State 2 And Synaptophysin Complex In Mouse Brain Isolated Synaptic Vesicles
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Intact V-Atpase State 3 And Synaptophysin Complex In Mouse Brain Isolated Synaptic Vesicles
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Intact V-Atpase State 3 In Synaptophysin Knock-Out Isolated Synaptic Vesicles
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Intact V-Atpase State 2 In Synaptophysin Knock-Out Isolated Synaptic Vesicles
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Intact V-Atpase State 1 And Synaptophysin Complex In Mouse Brain Isolated Synaptic Vesicles
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Intact V-Atpase State 1 In Synaptophysin Knock-Out Isolated Synaptic Vesicles
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
V0-Only V-Atpase In Synaptophysin Gene Knock-Out Mouse Brain Isolated Synaptic Vesicles
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: NAG, BMA |
 |
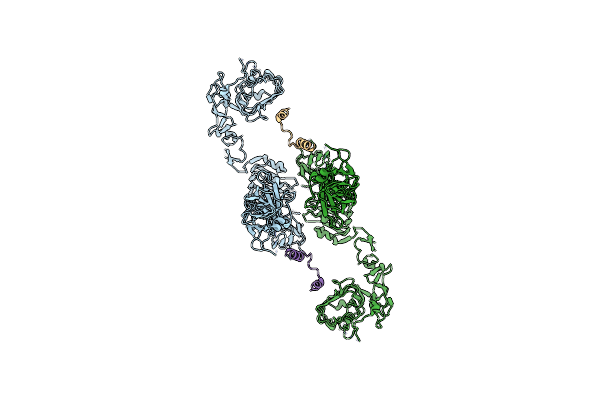
V0-Only V-Atpase And Synaptophysin Complex In Mouse Brain Isolated Synaptic Vesicles
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Organism: Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-07 Classification: SIGNALING PROTEIN |

