Search Count: 257
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE/INHIBITOR Ligands: PY1, CL |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE/INHIBITOR Ligands: CL, A1B7R, SO4 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: HOH |
 |
Crystal Structure Of The Immunity Protein (52 Domain-Containing Protein) From Pseudomonas Aeruginosa Pao1.
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: ANTITOXIN Ligands: EPE, PO4, EDO, CO3 |
 |
Crystal Structure Of T6Ss Effector-Immunity Complex Pa3907-Pa3908 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: UNKNOWN FUNCTION Ligands: CIT, MG |
 |
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: COA |
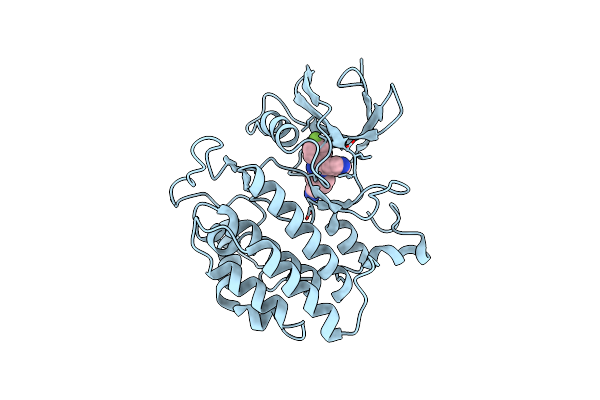 |
Crystal Structure Of Yck2 From Candida Albicans In Complex With Inhibitor 1E: 2-(4-Fluorophenyl)-3-(Pyridin-4-Yl)Imidazo[1,2-A]Pyridine-7-Carbonitrile
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-01-08 Classification: TRANSFERASE/INHIBITOR Ligands: A1BIQ, CL |
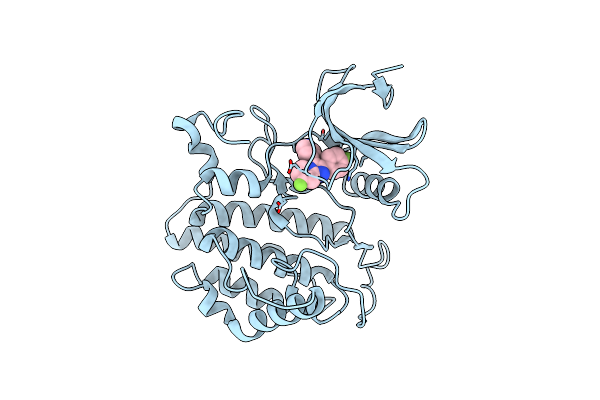 |
Crystal Structure Of Yck2 From Candida Albicans In Complex With Inhibitor 1F: 7-Fluoro-2-(4-Fluorophenyl)-3-(Pyridin-4-Yl)Imidazo[1,2-A]Pyridine
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-01-08 Classification: TRANSFERASE/INHIBITOR Ligands: A1BIP, CL |
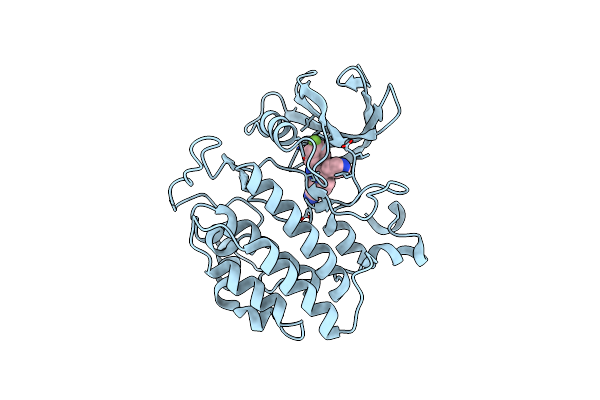 |
Crystal Structure Of Yck2 From Candida Albicans In Complex With Inhibitor 2A: 2-(4-Fluorophenyl)-3-(Pyridin-4-Yl)Pyrazolo[1,5-A]Pyridine-6-Carbonitrile
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-01-08 Classification: TRANSFERASE/INHIBITOR Ligands: A1BIO, CL |
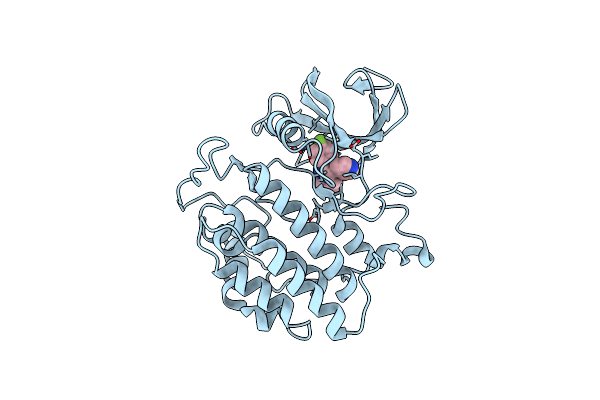 |
Crystal Structure Of Yck2 From Candida Albicans In Complex With Inhibitor 2B: 6-Fluoro-2-(4-Fluorophenyl)-3-(Pyridin-4-Yl)Pyrazolo[1,5-A]Pyridine
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-01-08 Classification: TRANSFERASE/INHIBITOR Ligands: A1BIN, CL |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SEY, PO4, CL |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SIN, EDO, PO4, CL |
 |
Organism: Clostridium carboxidivorans p7
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-03-13 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
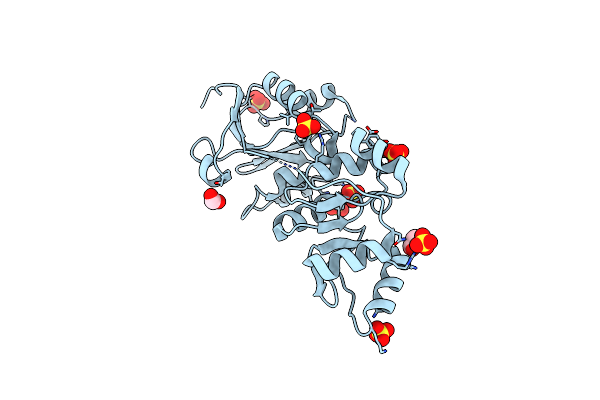
Crystal Structure Of Sars-Unique Domain (Sud) Of Nsp3 From Sars Coronavirus
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Release Date: 2023-10-18 Classification: VIRAL PROTEIN Ligands: CL, SO4 |
 |
Crystal Structure Of L516C/Y647C Mutant Of Sars-Unique Domain (Sud) From Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2023-10-18 Classification: VIRAL PROTEIN Ligands: FMT, ACT, SO4 |
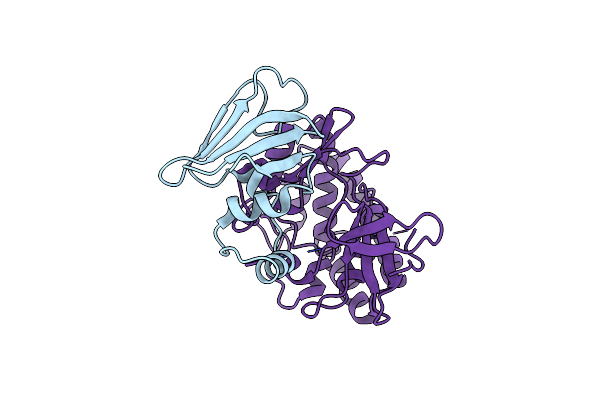 |
Crystal Structure Of The Hips(Lp)-Hipt(Lp) Complex From Legionella Pneumophila, Sel-Met Protein
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2023-09-27 Classification: TOXIN Ligands: CL |
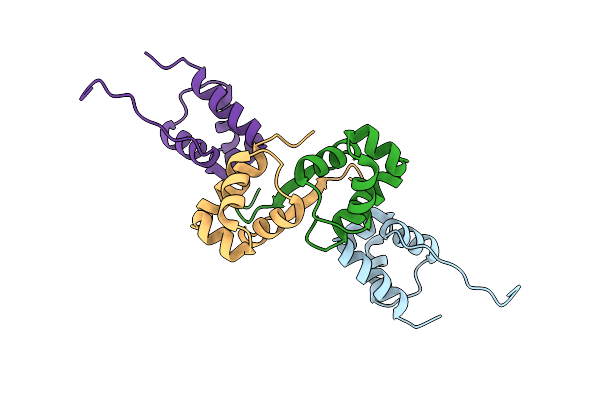 |
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-09-27 Classification: DNA BINDING PROTEIN Ligands: CL |
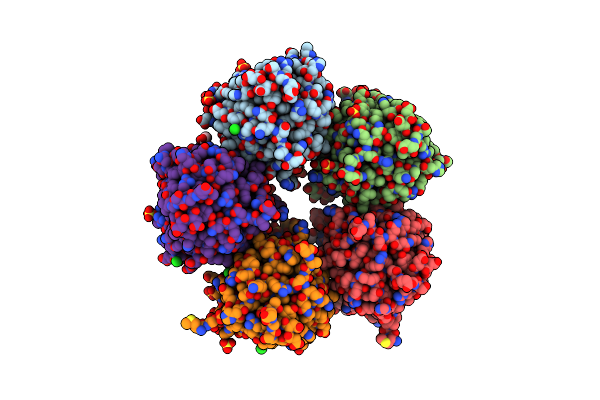 |
Receptor Shhtl5 From Striga Hermonthica In Complex With Strigolactone Agonist Gr24
Organism: Striga hermonthica
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2023-06-14 Classification: SIGNALING PROTEIN Ligands: TZU, SO4, EDO, CL |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul1 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR, GOL, SO4, CL |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: SO4, CL, GOL, PEG |

