Search Count: 23
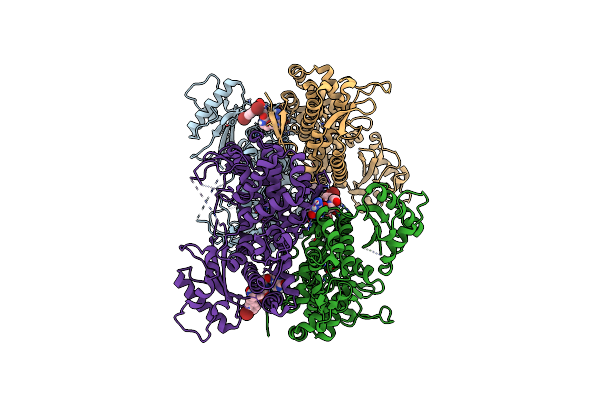 |
Crystal Structure Of Samhd1 Dimer Bound To An Inhibitor Obtained From High-Throughput Chemical Tethering To The Guanine Antiviral Acyclovir
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: A1BHL, FE |
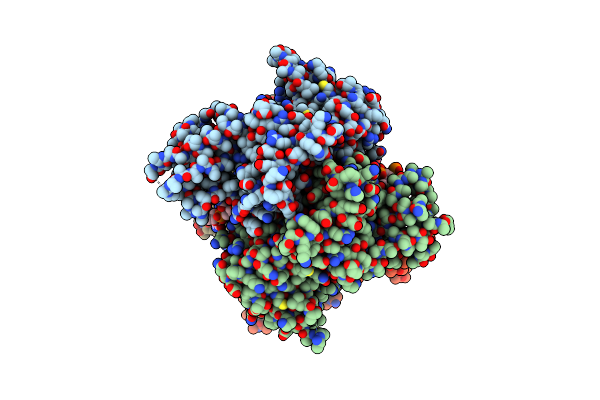 |
Organism: Homo sapiens, Virus-associated rnas
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: HYDROLASE Ligands: FE |
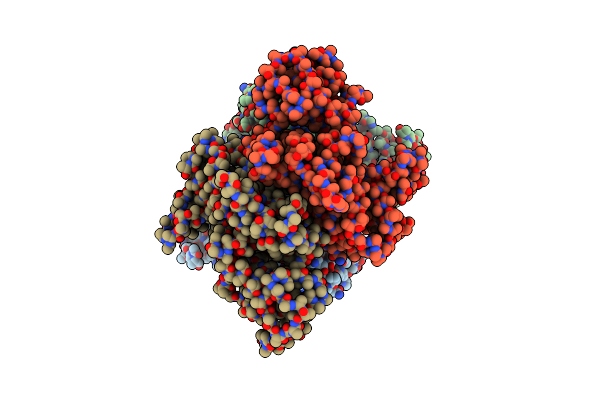 |
Organism: Homo sapiens, Virus-associated rnas
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: HYDROLASE Ligands: FE |
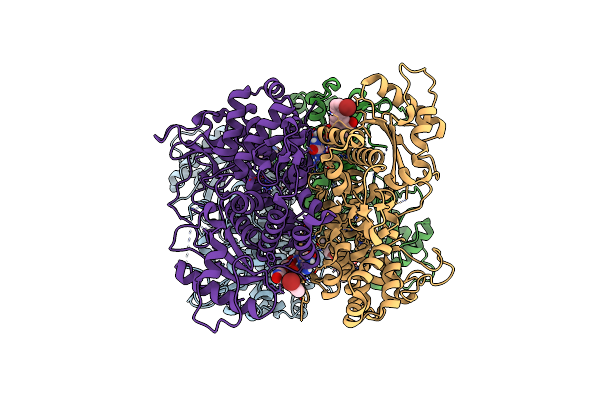 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: YWI, FE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: FE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: HYDROLASE Ligands: T8T |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2009-04-28 Classification: HYDROLASE Ligands: FCF, SCN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2009-04-28 Classification: HYDROLASE Ligands: 3FI, NA, SCN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2009-04-28 Classification: HYDROLASE Ligands: FCK |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-04-28 Classification: HYDROLASE Ligands: 3FL, SCN |
 |
Crystal Structure Of A Ung2/Modified Dna Complex That Represent A Stabilized Short-Lived Extrahelical State In Ezymatic Dna Base Flipping
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-10-30 Classification: Hydrolase/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-30 Classification: hydrolase/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: 302 |
 |
Uracil Dna Glycosylase Bound To A Cationic 1-Aza-2'-Deoxyribose-Containing Dna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-03-23 Classification: HYDROLASE/DNA Ligands: URA, PO4 |
 |
Solution Structure And Base Perturbation Studies Reveal A Novel Mode Of Alkylated Base Recognition By 3-Methyladenine Dna Glycosylase I
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2003-11-25 Classification: HYDROLASE Ligands: ZN, ADK |
 |
Nmr Solution Structure Of Zinc-Binding Protein 3-Methyladenine Dna Glycosylase I (Tag)
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2003-06-03 Classification: HYDROLASE Ligands: ZN |
 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2001-01-17 Classification: HYDROLASE Ligands: URA |
 |
Crystal Structure Of Escherichia Coli Uracil Dna Glycosylase And Its Complexes With Uracil And Glycerol: Structure And Glycosylase Mechanism Revisited
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 1999-10-13 Classification: HYDROLASE Ligands: URA |
 |
Crystal Structure Of Escherichia Coli Uracil Dna Glycosylase And Its Complexes With Uracil And Glycerol: Structure And Glycosylase Mechanism Revisited
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 1999-10-13 Classification: HYDROLASE Ligands: GOL |

