Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
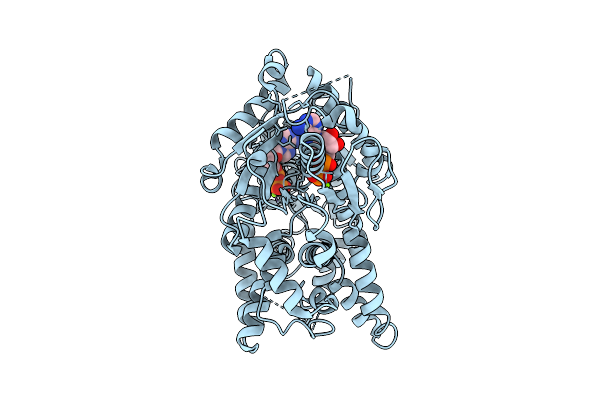 |
Open Conformation Of Arsa From L. Ferriphilum In Complex With Mgadp Determined In The Presence Of Arsenite
Organism: Leptospirillum ferriphilum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE Ligands: ADP, MG |
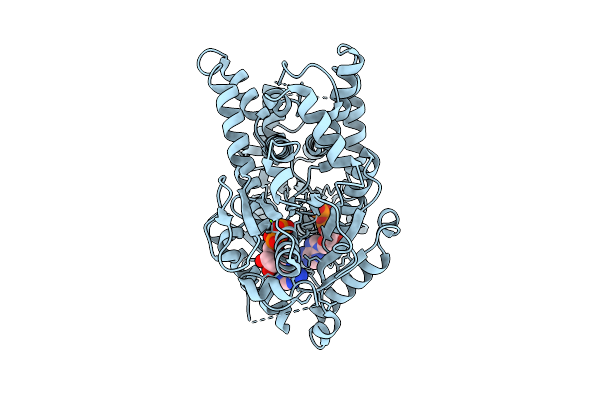 |
Open Conformation Of Arsa From L. Ferriphilum In Complex With Mgadp Determined In Absence Of Arsenite
Organism: Leptospirillum ferriphilum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE Ligands: ADP, MG |
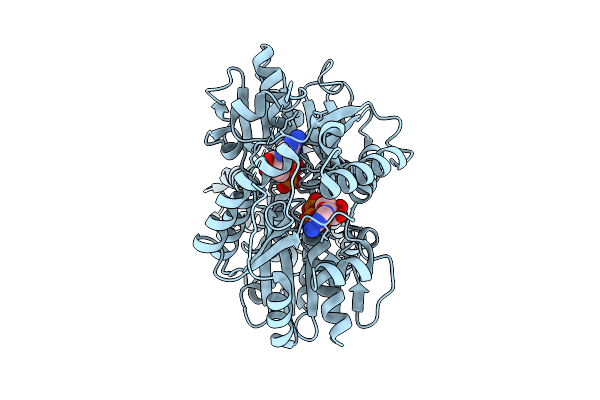 |
Closed Conformation Of Arsa From L. Ferriphilum In Complex With Mgatp And Arsenite
Organism: Leptospirillum ferriphilum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE Ligands: ATP, MG, ARS |
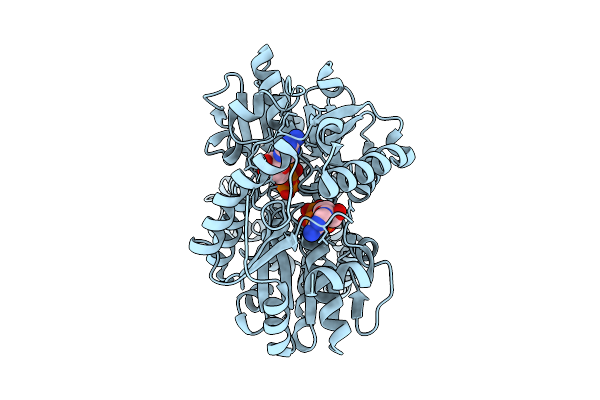 |
Closed Conformation Of Arsa From L. Ferriphilum In Complex With Mgatp And Arsenite At 1.5 Minute Time Point
Organism: Leptospirillum ferriphilum ml-04
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE Ligands: ATP, MG, ARS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: TRANSFERASE Ligands: SAH, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-15 Classification: TRANSFERASE Ligands: GOL, SAH, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-01-15 Classification: TRANSFERASE Ligands: SAH, ZN, PEG, PG4, EOH, GOL |
 |
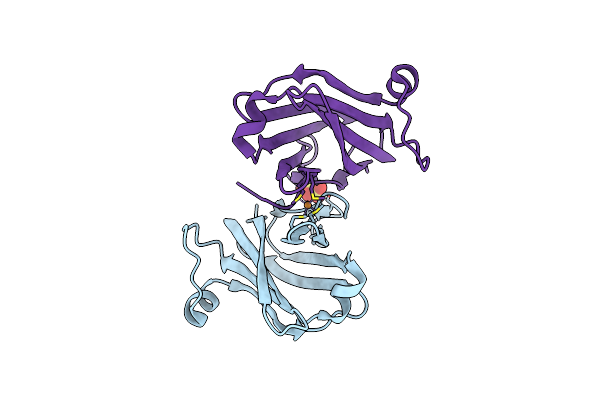
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-06-22 Classification: METAL TRANSPORT, Oxidoreductase Ligands: SO4 |
 |
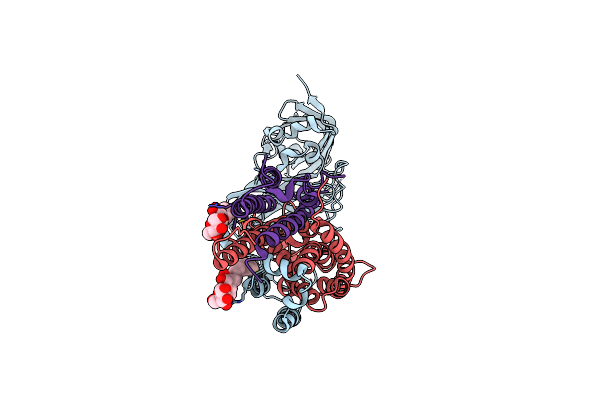
Crystal Structure Of Pmod Soluble Domain From Methylocystis Sp. Atcc 49242 (Rockwell)
Organism: Methylocystis sp. atcc 49242
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-10-31 Classification: METAL BINDING PROTEIN Ligands: SO4, CU1 |
 |
Crystal Structure Of Particulate Methane Monooxygenase From Methylomicrobium Alcaliphilum 20Z
Organism: Methylomicrobium alcaliphilum 20z
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: CU, CM5 |
 |
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2015-07-08 Classification: HYDROLASE Ligands: CD, TCE |
 |
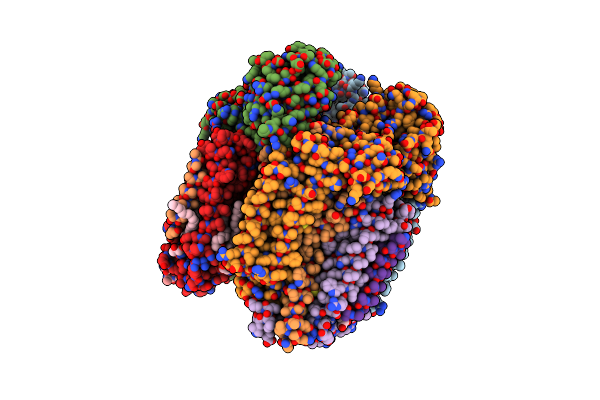
Solution Structure Of A De Novo Designed Peptide That Sequesters Toxic Heavy Metals
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2015-04-01 Classification: DE NOVO PROTEIN |
 |
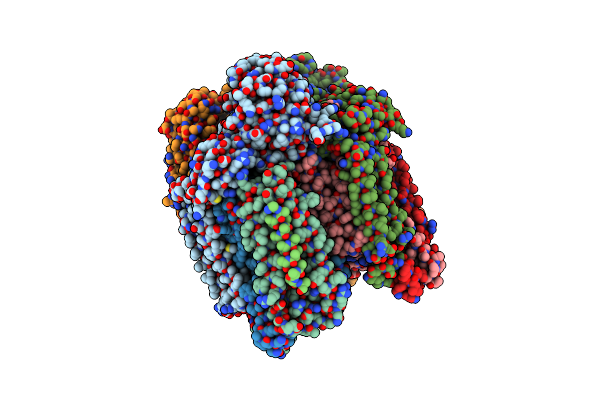
Crystal Structure Of Particulate Methane Monooxygenase From Methylocystis Sp. Atcc 49242 (Rockwell)
Organism: Methylocystis sp. atcc 49242
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-06-25 Classification: OXIDOREDUCTASE Ligands: CU, PGT |
 |
Crystal Structure Of Particulate Methane Monooxygenase From Methylocystis Sp. Atcc 49242 (Rockwell) Soaked In Copper
Organism: Methylocystis sp. atcc 49242
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2014-06-25 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Crystal Structure Of Particulate Methane Monooxygenase From Methylocystis Sp. Atcc 49242 (Rockwell) Soaked In Zinc
Organism: Methylocystis sp. atcc 49242
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2014-06-25 Classification: OXIDOREDUCTASE Ligands: CU, ZN, CAC |
 |
Crystal Structure Of Particulate Methane Monooxygenase (Pmmo) From Methylocystis Sp. Strain M
Organism: Methylocystis sp. m
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2011-11-23 Classification: OXIDOREDUCTASE Ligands: CUA, ZN, CU |
 |
Crystal Structure Of Particulate Methane Monooxygenase From Methylococcus Capsulatus (Bath)
Organism: Methylococcus capsulatus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-11-23 Classification: OXIDOREDUCTASE Ligands: CU, ZN, CUA |
 |
Crystal Structure Of The Staphylococcus Aureus Pi258 Cadc Metal Binding Site 2 Mutant
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2009-04-07 Classification: DNA BINDING PROTEIN, GENE REGULATION |
 |
Crystal Structure Of Methylosinus Trichosporium Ob3B Particulate Methane Monooxygenase (Pmmo)
Organism: Methylosinus trichosporium
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2008-07-15 Classification: MEMBRANE PROTEIN Ligands: CU |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2007-07-03 Classification: METAL TRANSPORT Ligands: ZN, FES, ACY |