Search Count: 20
 |
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-02-15 Classification: DNA BINDING PROTEIN |
 |
Structure Of The Toll - Spatzle Complex, A Molecular Hub In Drosophila Development And Innate Immunity
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-04-09 Classification: IMMUNE SYSTEM/CYTOKINE Ligands: NAG, EPE |
 |
Structure Of The Toll - Spatzle Complex, A Molecular Hub In Drosophila Development And Innate Immunity (Glycosylated Form)
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2014-04-09 Classification: IMMUNE SYSTEM/CYTOKINE Ligands: NAG, EPE |
 |
Crystal Structure Of Full Length Deinococcus Radiodurans Uvrd In Complex With Dna
Organism: Deinococcus radiodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2013-10-30 Classification: HYDROLASE/DNA Ligands: MG, ANP |
 |
Crystal Structure Of Deinococcus Radiodurans Uvrd In Complex With Dna, Form 1
Organism: Deinococcus radiodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2013-10-30 Classification: HYDROLASE/DNA Ligands: ANP, MG, NO3, GOL, NA |
 |
Crystal Structure Of Deinococcus Radiodurans Uvrd In Complex With Dna, Form 2
Organism: Deinococcus radiodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-10-30 Classification: HYDROLASE/DNA Ligands: ANP, MG |
 |
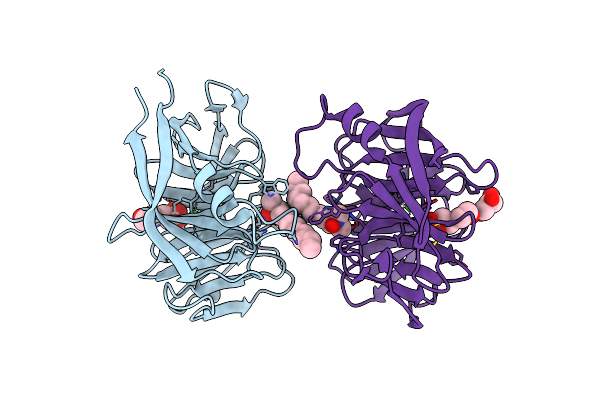
Crystal Structure Of C-Terminal Domain Of Helicobacter Pylori Dnab Helicase
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-03-21 Classification: HYDROLASE Ligands: FLC |
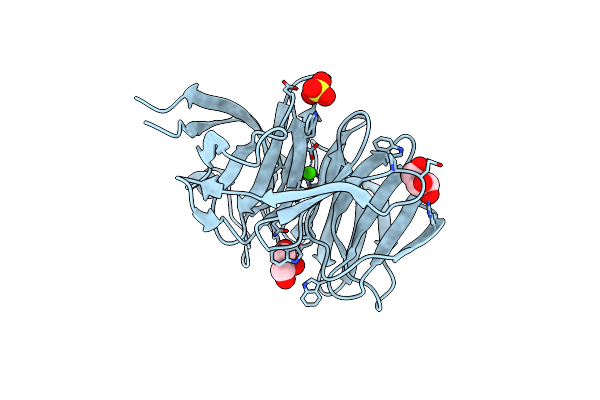
 |
Organism: Myxococcus xanthus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2010-11-03 Classification: TRANSFERASE Ligands: CA, MES, DDQ, GOL |
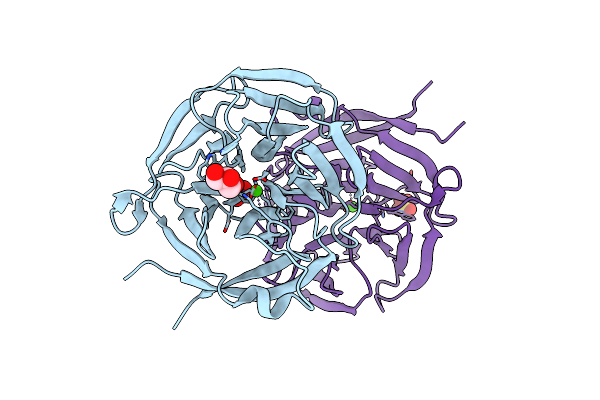
 |
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-11-03 Classification: TRANSFERASE Ligands: CA, GOL, SO4 |
 |
Crystal Structure Of Zymomonas Mobilis Glutaminyl Cyclase (Monoclinic Form)
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-11-03 Classification: TRANSFERASE Ligands: CA, GOL, SO4 |
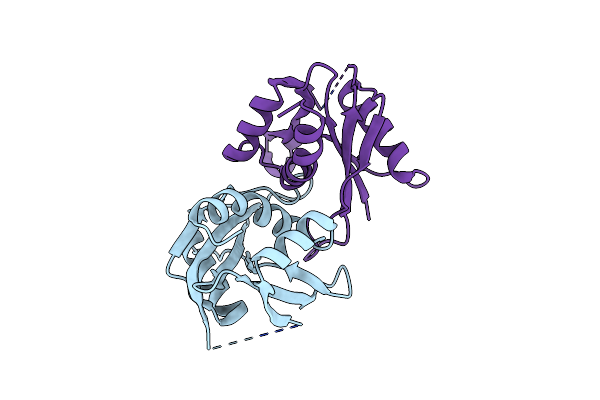
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2009-12-15 Classification: ELECTRON TRANSPORT Ligands: SF4 |
 |
Structure Of An N-Terminally Truncated Nudix Hydrolase Dr2204 From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-12-01 Classification: HYDROLASE |
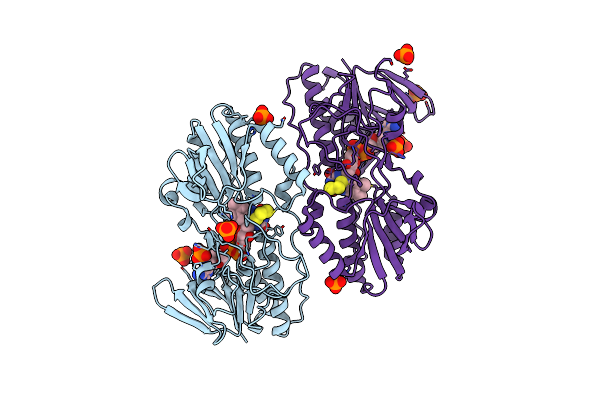 |
The First X-Ray Structure Of A Sulfide:Quinone Oxidoreductase: Insights Into Sulfide Oxidation Mechanism
Organism: Acidianus ambivalens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2009-06-02 Classification: OXIDOREDUCTASE Ligands: FAD, S3H, PO4 |
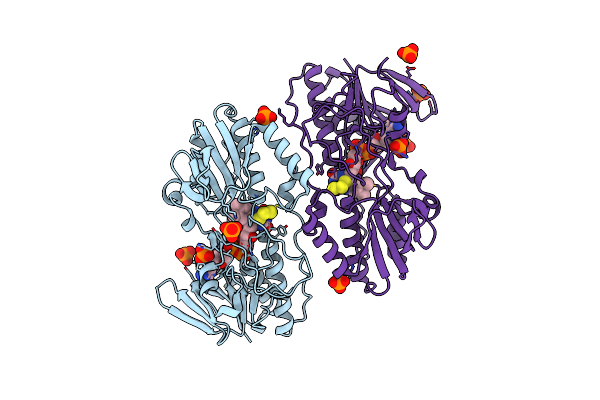 |
The First X-Ray Structure Of A Sulfide:Quinone Oxidoreductase: Insights Into Sulfide Oxidation Mechanism
Organism: Acidianus ambivalens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2009-06-02 Classification: OXIDOREDUCTASE Ligands: FAD, S3H, PO4 |
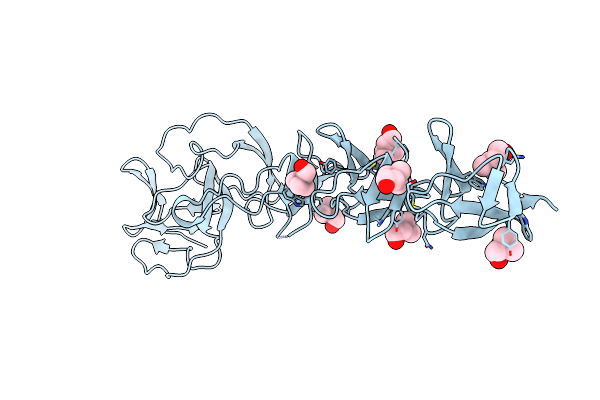 |
Crystal Structure Of Choline Binding Protein F From Streptococcus Pneumoniae In The Presence Of A Peptidoglycan Analogue (Tetrasaccharide-Pentapeptide)
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2009-02-03 Classification: CHOLINE-BINDING PROTEIN Ligands: CHT |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2008-10-28 Classification: ELECTRON TRANSPORT Ligands: SO4, HEC |
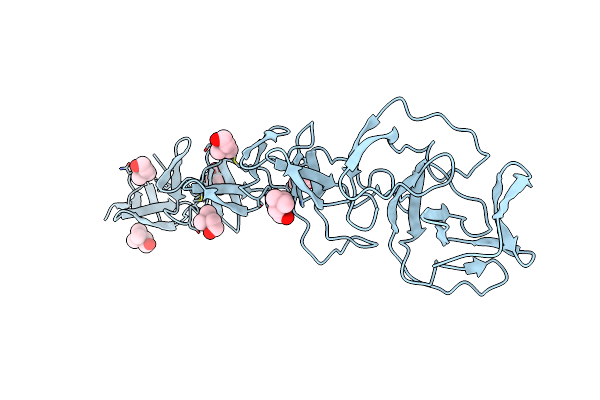 |
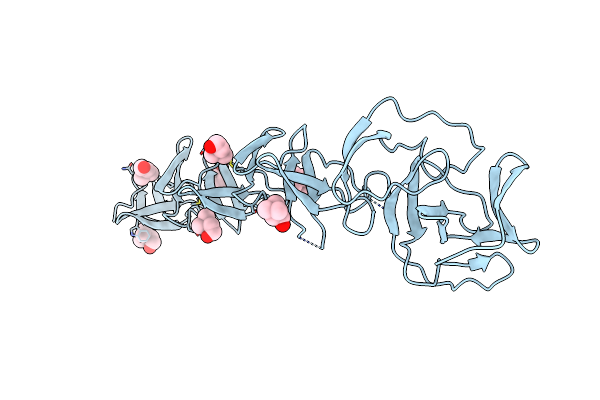
Crystal Structure Of Choline Binding Protein F From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-06-10 Classification: LIPID BINDING PROTEIN Ligands: CHT |
 |
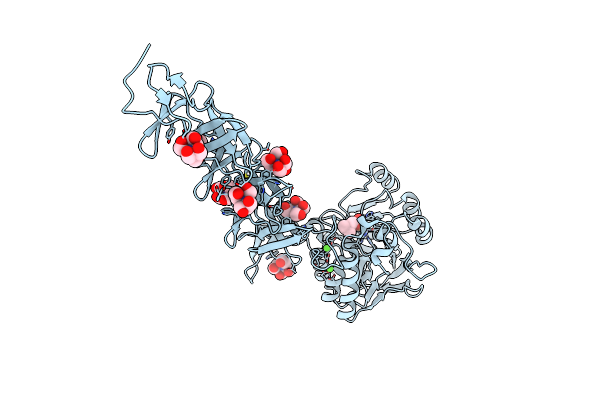
Crystal Structure Of Choline Binding Protein F From Streptococcus Pneumoniae. Crystal Form Ii.
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2008-06-10 Classification: LIPID BINDING PROTEIN Ligands: CHT |
 |
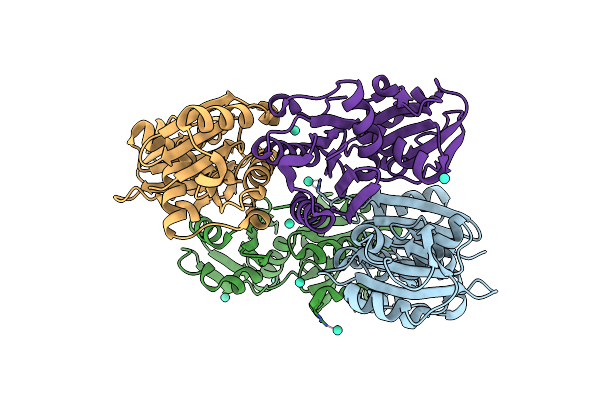
Crystal Structure Of The Complete Modular Teichioic Acid Phosphorylcholine Esterase Pce (Cbpe) From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2005-05-09 Classification: HYDROLASE Ligands: PC, BTB, ZN, CA |
 |
Crystal Structure Of The Essential E. Coli Yeaz Protein By Mad Method Using The Gadolinium Complex "Dotma"
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2004-09-16 Classification: HYDROLASE Ligands: GD3 |

