Search Count: 19
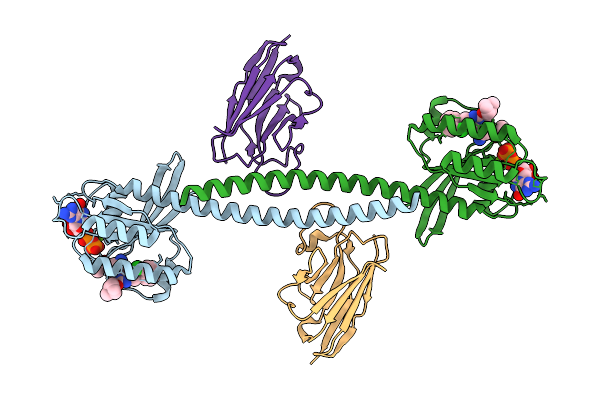 |
A Coiled Coil Module Strategy For High-Resolution Cryo-Em Structures Of Small Proteins For Drug Discovery
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: STRUCTURAL PROTEIN Ligands: M1X, GDP, MG |
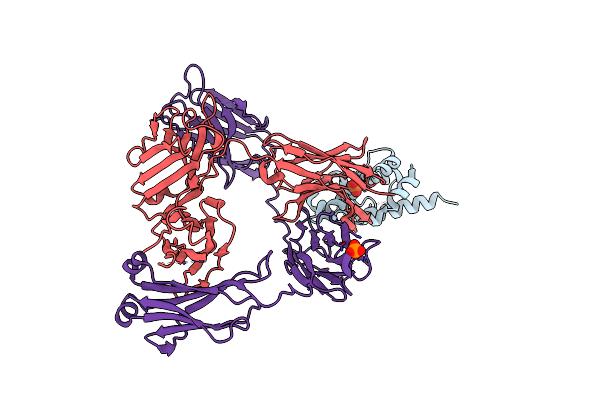 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-03-30 Classification: IMMUNE SYSTEM Ligands: PO4 |
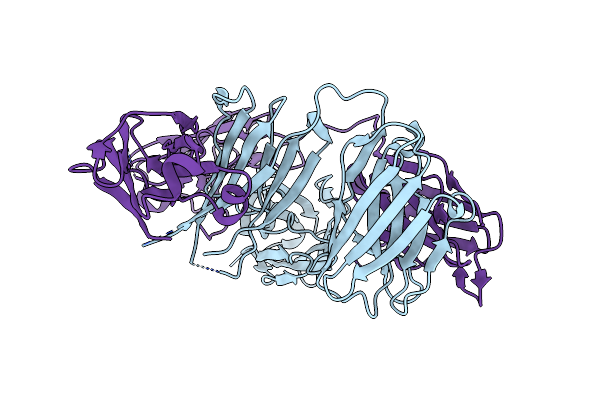 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2016-03-30 Classification: IMMUNE SYSTEM |
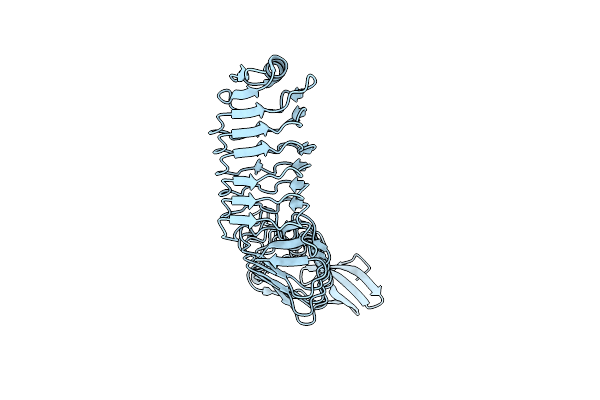 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2015-02-25 Classification: CHOLINE-BINDING PROTEIN |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION, EPR Resolution:1.64 Å Release Date: 2013-07-31 Classification: CHAPERONE Ligands: GOL |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION, EPR Resolution:2.90 Å Release Date: 2013-07-31 Classification: CHAPERONE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-01-01 Classification: LYASE Ligands: MG, FAD, TPP, DTT, 1PE |
 |
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2006-04-25 Classification: OXIDOREDUCTASE Ligands: MG, PO4, NA, TPP, FAD |
 |
Pyruvate Oxidase Variant F479W In Complex With Reaction Intermediate 2-Lactyl-Thiamin Diphosphate
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2006-04-25 Classification: OXIDOREDUCTASE Ligands: MG, NA, TDL, FAD, PYR |
 |
Pyruvate Oxidase Variant F479W In Complex With Reaction Intermediate Analogue 2-Phosphonolactyl-Thiamin Diphosphate
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-04-25 Classification: OXIDOREDUCTASE Ligands: MG, NA, TDK, FAD |
 |
Pyruvate Oxidase Variant F479W In Complex With Reaction Intermediate 2-Hydroxyethyl-Thiamin Diphosphate
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2006-04-25 Classification: OXIDOREDUCTASE Ligands: MG, PO4, NA, TDM, FAD, PYR |
 |
Pyruvate Oxidase Variant F479W In Complex With Reaction Intermediate 2-Acetyl-Thiamin Diphosphate
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2006-04-25 Classification: OXIDOREDUCTASE Ligands: MG, NA, HTL, FAD, PYR |
 |
X-Ray Structure Of The Orphan Nuclear Receptor Ror Beta Ligand-Binding Domain In The Active Conformation
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-04-09 Classification: HORMONE/GROWTH FACTOR Ligands: STE |
 |
Nature Of The Inactivation Of Elastase By N-Peptidyl-O-Aroyl Hydroxylamine As A Function Of Ph
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-07-10 Classification: COMPLEX (HYDROLASE/INHIBITOR) Ligands: CA, SO4, BAF |
 |
Nature Of The Inactivation Of Elastase By N-Peptidyl-O-Aroyl Hydroxylamine As A Function Of Ph
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 1995-07-10 Classification: COMPLEX (HYDROLASE/INHIBITOR) Ligands: CA, BAA |
 |
Three-Dimensional Structure Of Chymotrypsin Inactivated With (2S) N-Acetyl-L-Alanyl-L-Phenylalanyl-Chloroethyl Ketone: Implications For The Mechanism Of Inactivation Of Serine Proteases By Chloroketones
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-11-01 Classification: HYDROLASE(SERINE PROTEINASE) Ligands: HIN |
 |
Inactivation Of Subtilisin Carlsberg By N-(Tert-Butoxycarbonyl-Alanyl-Prolyl-Phenylalanyl)-O-Benzol Hydroxylamine: Formation Of Covalent Enzyme-Inhibitor Linkage In The Form Of A Carbamate Derivative
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1994-08-31 Classification: hydrolase/hydrolase INHIBITOR Ligands: 0EF, CA, NA |
 |
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1994-01-31 Classification: SERINE PROTEASE Ligands: CA, NA |
 |
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1994-01-31 Classification: SERINE PROTEASE Ligands: CA, CCN |

