Search Count: 81
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: A1JNK, ADP |
 |
Dye Type Peroxidase Aa From Streptomyces Lividans With N3 Ligand By Serial Electron Diffraction (Serialed)
Organism: Streptomyces lividans
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: AZI, HEM |
 |
Dye Type Peroxidase Aa From Streptomyces Lividans By Microcrystal Electron Diffraction (Microed/3D Ed)
Organism: Streptomyces lividans
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Dye Type Peroxidase Aa From Streptomyces Lividans By Serial Electron Diffraction (Serialed)
Organism: Streptomyces lividans
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-01-22 Classification: CYTOKINE Ligands: A1ISH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: CYTOKINE Ligands: SO4, A1ISG |
 |
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-14-14 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-01-15 Classification: DNA BINDING PROTEIN Ligands: A1H1R, A1H1Q, DTV |
 |
Microtubule-Associated Kinesin-1 Tail Complex Bound To Amppnp, Single-Headed State
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: MOTOR PROTEIN Ligands: GDP, TA1, ANP, MG, GTP |
 |
Microtubule-Associated Kinesin-1 Tail Complex Bound To Amppnp, Two-Headed State
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: MOTOR PROTEIN Ligands: GDP, TA1, ANP, MG, GTP |
 |
Microtubule-Associated Kinesin-1 Tail Complex Bound To Adp, Single-Headed State
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: MOTOR PROTEIN Ligands: GDP, TA1, GTP, MG, ADP |
 |
Microtubule-Associated Kinesin-1 Tail Complex Bound To Adp, Two-Headed State
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: MOTOR PROTEIN Ligands: GDP, TA1, GTP, MG, ADP |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: MOTOR PROTEIN Ligands: GDP, TA1, GTP, MG, ADP |
 |
Crystal Structure Of The Cysteine-Rich Gallus Gallus Urate Oxidase In Complex With The 8-Azaxanthine Inhibitor Under Reducing Conditions (Space Group C 2 2 21)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, EDO, CL |
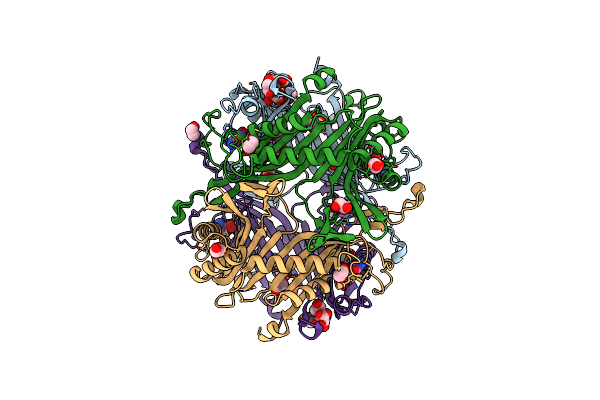 |
Crystal Structure Of The Cysteine-Rich Gallus Gallus Urate Oxidase In Complex With The 8-Azaxanthine Inhibitor Under Reducing Conditions (Space Group P 21 21 21)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: AZA, EDO, TAR, OXY, CL |
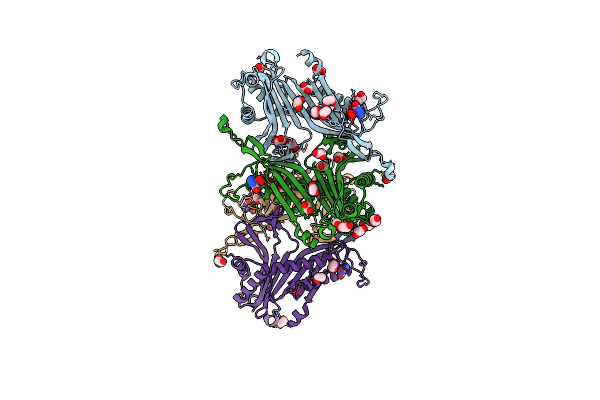 |
Crystal Structure Of The Cysteine-Rich Gallus Gallus Urate Oxidase In Complex With The 8-Azaxanthine Inhibitor Under Oxidising Conditions (Space Group C 2 2 21)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, EDO, CL, BR |
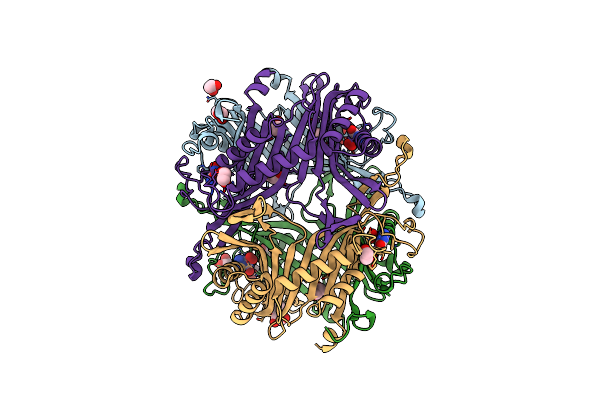 |
Crystal Structure Of The Cysteine-Rich Gallus Gallus Urate Oxidase In Complex With The 8-Azaxanthine Inhibitor Under Oxidising Conditions (Space Group P 21 21 21)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, EDO, CL |
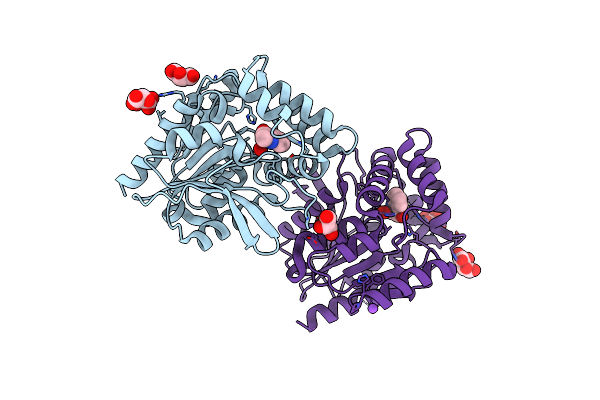 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) S101A Variant Complexed With 2-Methyl-Quinolin-4(1H)-One Under Hyperoxyc Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: VFH, OXY, TAR, GOL, NA |
 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) S101A Variant Complexed With 2-Methyl-Quinolin-4(1H)-One Under Normoxyc Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: VFH, GOL, SRT |
 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) H251A Variant Complexed With N-Acetylanthranilate As Result Of In Crystallo Turnover Of Its Natural Substrate 1-H-3-Hydroxy-4- Oxoquinaldine Under Hyperoxic Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: ZZ8, SRT, GOL, K |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: NEUTRON DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: METAL BINDING PROTEIN Ligands: FE |

