Search Count: 24
 |
Crystal Structure Of A Manganese-Containing Cupin (Tm1459) From Thermotoga Maritima, Variant C106Q
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN Ligands: CL |
 |
Crystal Structure Of A Manganese-Containing Cupin (Tm1459) From Thermotoga Maritima, Variant Aifq (Y7A/M38I/Y83F/C106Q)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN |
 |
Crystal Structure Of A Manganese-Containing Cupin (Tm1459) From Thermotoga Maritima, Variant 208 (V19I/R23H/M38I/I60F/C106Q)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN |
 |
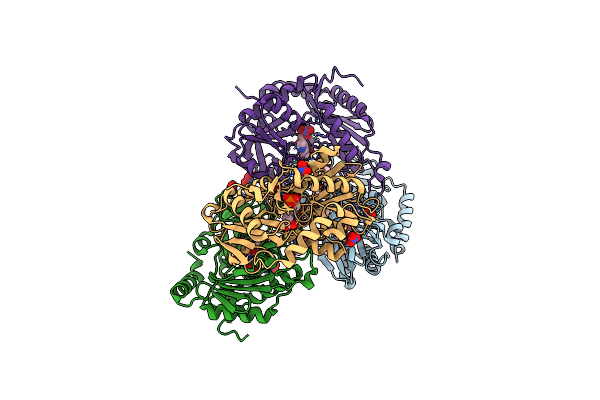
Organism: Exophiala sideris
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2020-12-09 Classification: TRANSFERASE Ligands: PLP, GOL, CL, P6G |
 |
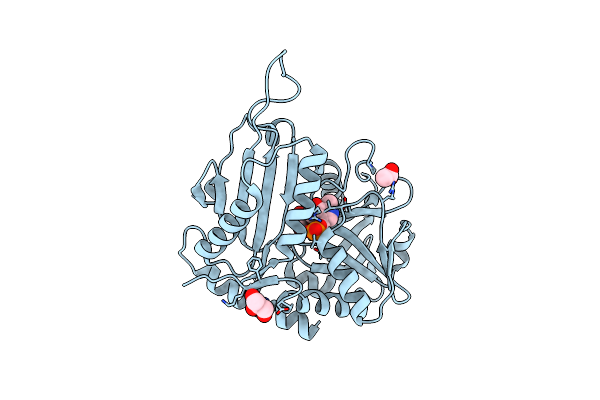
Organism: Shinella sp. jr1-6
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-09 Classification: TRANSFERASE Ligands: PLP, CL, GOL, NO3, PEG, PLG, GAB |
 |
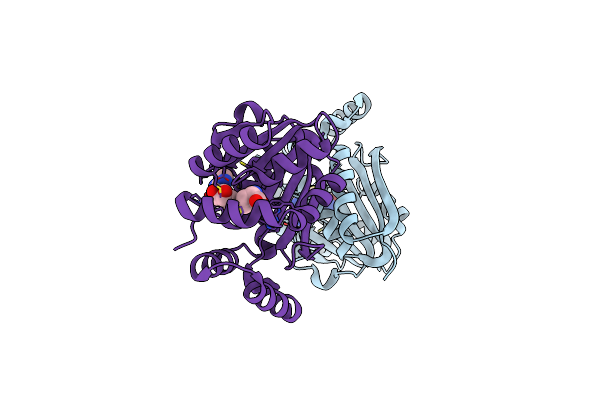
Crystal Structure Of An (R)-Selective Amine Transaminase From Exophiala Xenobiotica
Organism: Exophiala xenobiotica
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2019-01-09 Classification: BIOSYNTHETIC PROTEIN Ligands: PLP, GOL, ACT |
 |
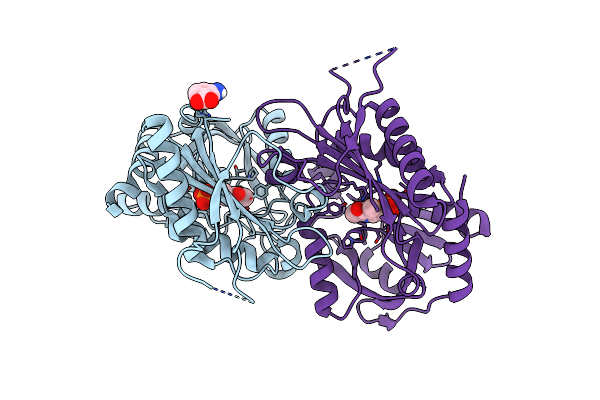
Crystal Structure Of Couo, A C-Methyltransferase From Streptomyces Rishiriensis
Organism: Streptomyces rishiriensis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-02-15 Classification: TRANSFERASE Ligands: SAH |
 |
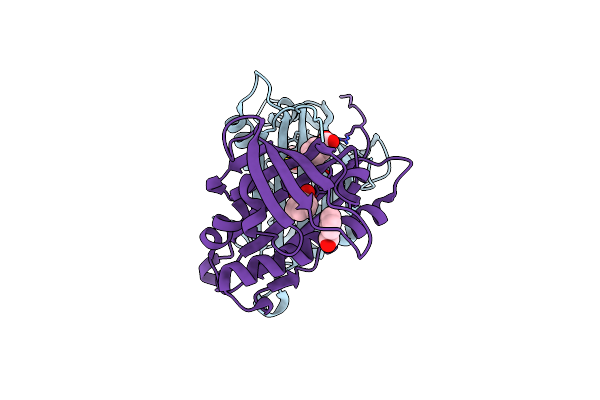
Structural Characterisation Of Fold Iv-Transaminase, Cputa1, From Curtobacterium Pusillum
Organism: Curtobacterium pusillum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-12-14 Classification: TRANSFERASE Ligands: PLP, GAB |
 |
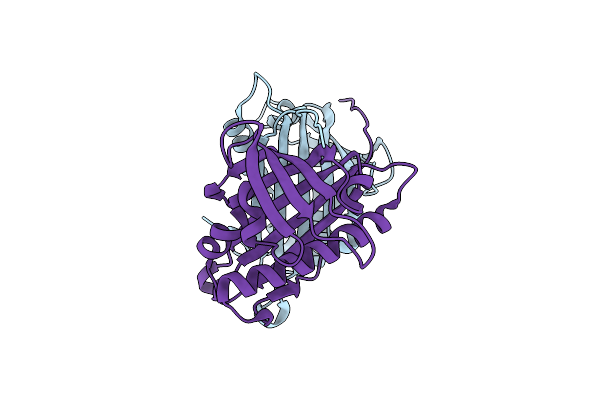
Hydroxynitrile Lyase From The Fern Davallia Tyermanii In Complex With Benzoic Acid
Organism: Davallia tyermannii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-10-12 Classification: LYASE Ligands: BEZ |
 |
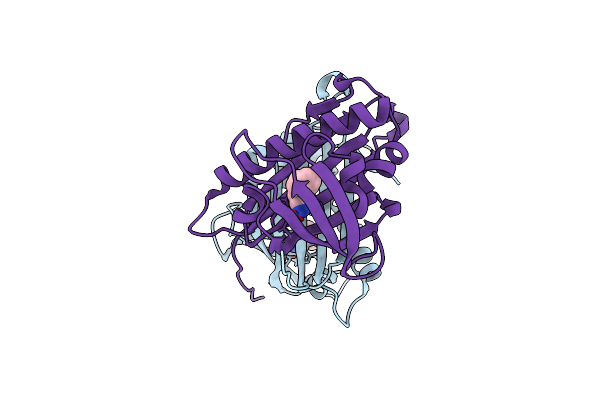
Organism: Davallia tyermannii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-10-05 Classification: LYASE |
 |
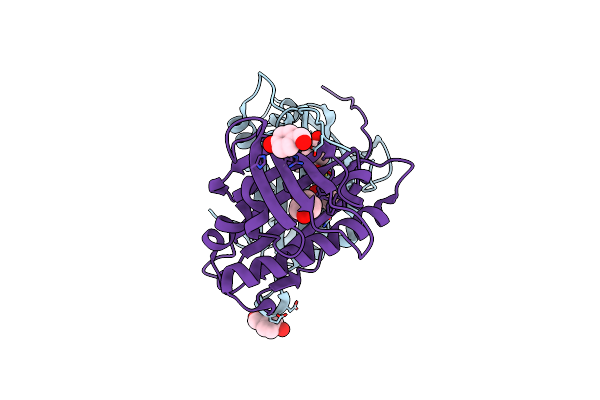
Hydroxynitrile Lyase From The Fern Davallia Tyermanii In Complex With (R)-Mandelonitrile / Benzaldehyde
Organism: Davallia tyermannii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-10-05 Classification: LYASE Ligands: HBX, MXN |
 |
Hydroxynitrile Lyase From The Fern Davallia Tyermanii In Complex With P-Hydroxybenzaldehyde
Organism: Davallia tyermannii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-05 Classification: LYASE Ligands: HBA |
 |
Improved Variant Of (R)-Selective Manganese-Dependent Hydroxynitrile Lyase From Bacteria
Organism: Granulicella tundricola
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-01-21 Classification: LYASE Ligands: MN |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound P-Hydroxybenzaldehyde
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HBA |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound Benzene-1,4-Diol
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HQE |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound P-Hydroxybenzaldehyde
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HBA, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound Benzene-1,4-Diol
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HQE, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound 1-Cyclohex-2-Enone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: FMN, A2Q |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound 1-Cyclohex-2-Enone
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: A2Q, FMN |
 |
First Crystal Structure Of An (R)-Selective Omega-Transaminase From Aspergillus Terreus
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2014-02-12 Classification: TRANSFERASE Ligands: PLP, PDG, CL, CA |

