Search Count: 281
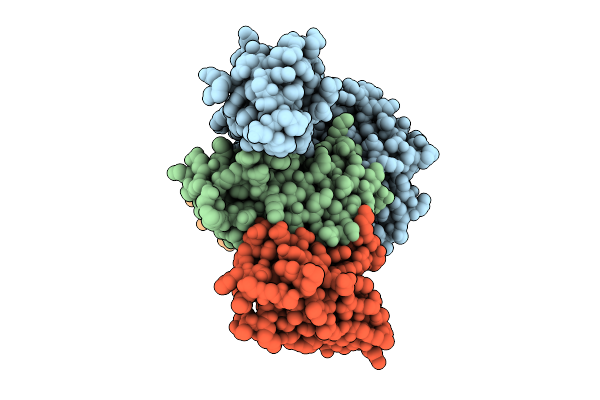 |
Organism: Homo sapiens, Lama glama, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Rabbit hemorrhagic disease virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: EDO, NHE, TRS, PEG |
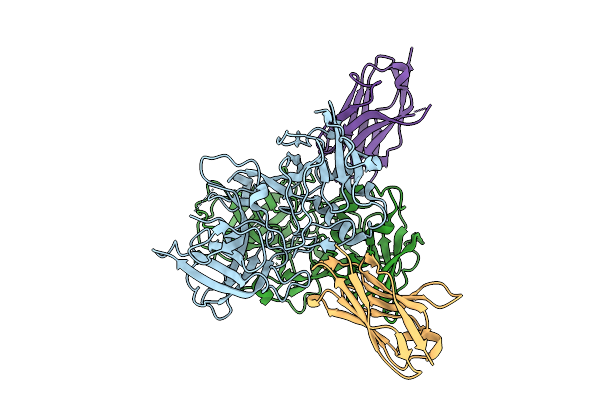 |
Organism: Hare calicivirus, Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN |
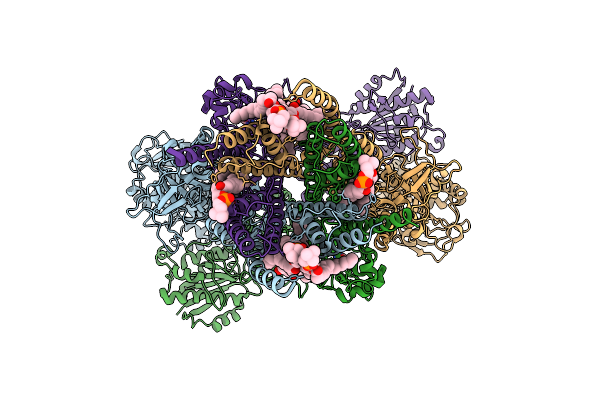 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: GLY, POV, GLU, A1CE1 |
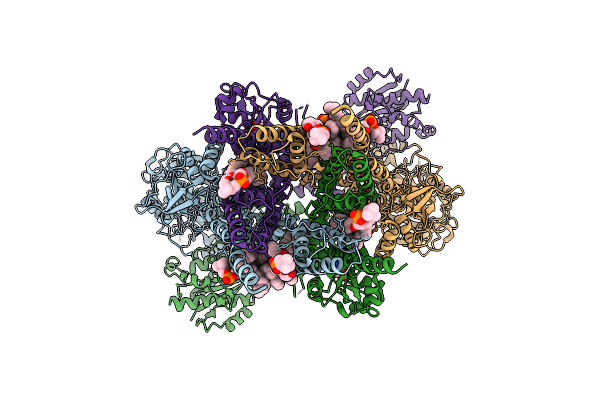 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: GLY, POV, GLU, A1CE1 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: GLY, POV, GLU, A8W |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: GLY, POV, GLU, A8W |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: POV, A1AFT, CLR |
 |
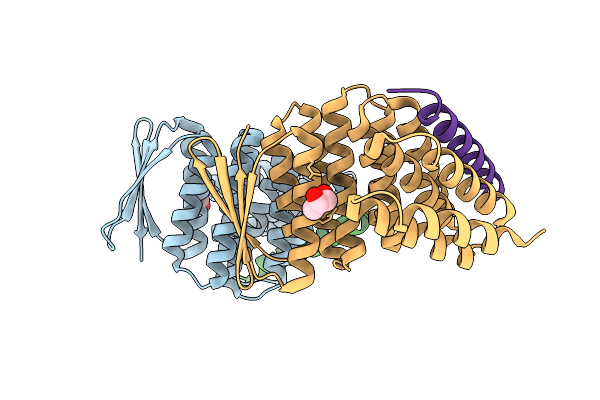
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: CL |
 |
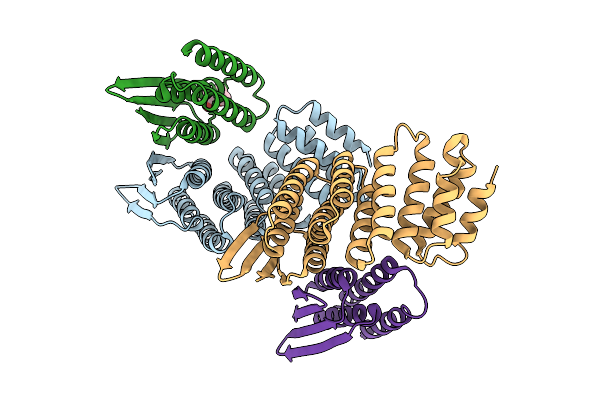
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: PGO |
 |
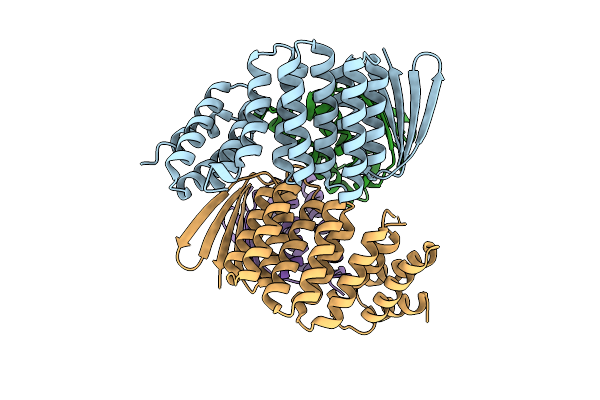
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: MPD |
 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
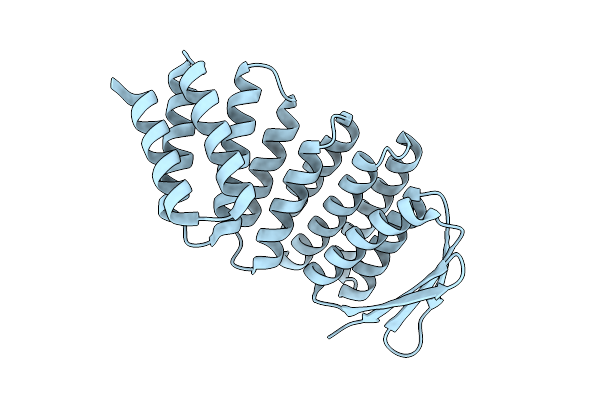
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State The With Methylated Lysines
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Facilitated Dissociation Target Lhd101An1: Lhd101A With An N-Terminal Extension
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Conformational Switch Effector Peptide Cs221B
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Designed Allosteric Facilitated Dissociation Switch As1_K46L_E50W_K172W_E173Y In Complex State The
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of The Sars-Cov-2 Helicase Nsp13 In Complex With Myricetin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: ZN, PO4, MPO, MYC |

