Search Count: 256
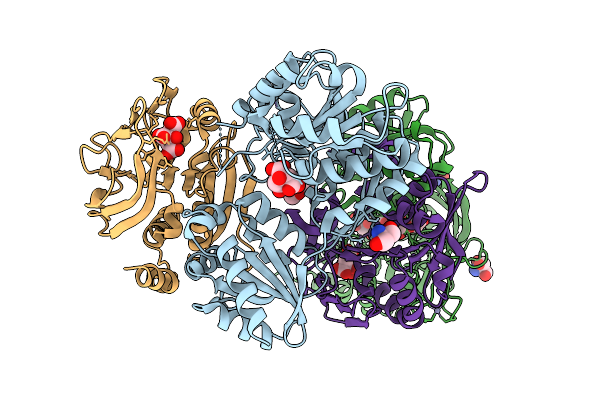 |
Crystal Structure Of The Murt/Gatd Enzyme Complex From Streptococcus Pyogenes
Organism: Streptococcus pyogenes mgas10270
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIGASE Ligands: ZN, CIT, TRS, PEG |
 |
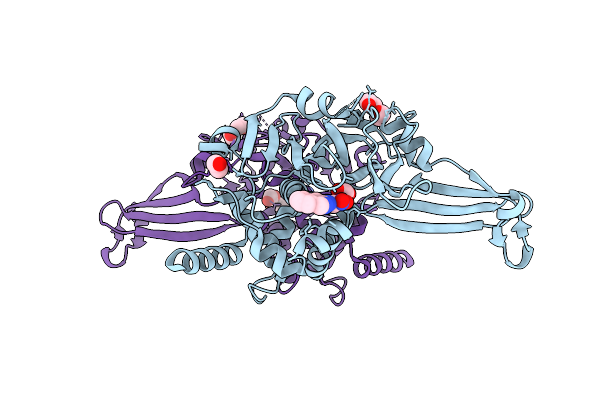
Crystal Structure Of The Murt/Gatd Enzyme Complex From Streptococcus Pyogenes With Bound Amppnp
Organism: Streptococcus pyogenes mgas10270
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIGASE Ligands: ZN, MG, ANP, GOL |
 |
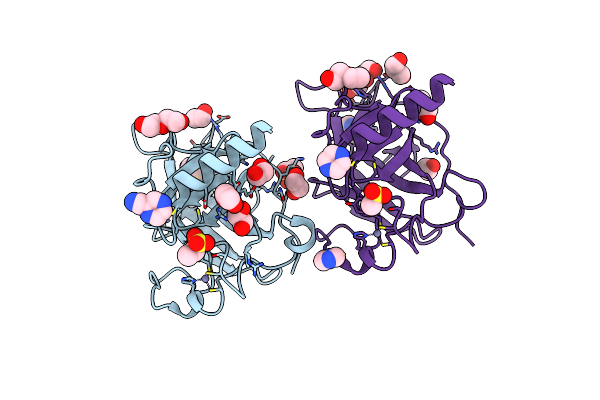
Usp7 Covalently Bound To N-(6-Fluoro-3-Nitropyridin-2-Yl)-5-(1-Methyl-1H-Pyrazol-4-Yl)Isoquinolin-3-Amine (Gcl36, 7A) With Partial Occupancy
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: A1I71, EDO, PEG, BR |
 |
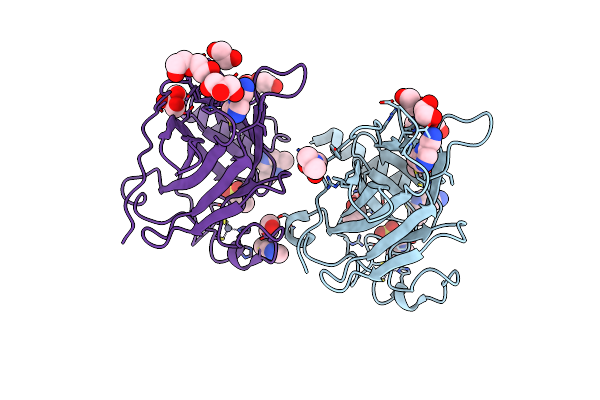
P53-Y220C Core Domain Covalently Bound To 2-Chloro-5-Cyanopyrazine Soaked At 5 Mm
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: CELL CYCLE Ligands: A1IIO, EPE, PEG, EDO, ZN, GOL |
 |
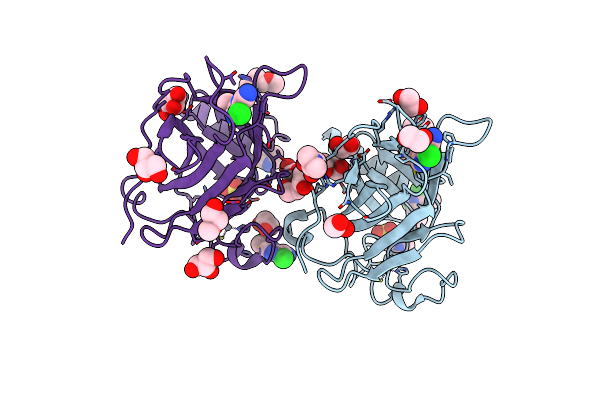
P53-Y220C Core Domain Covalently Bound To 5-Chloro-6-Methylpyrazine-2-Carbonitrile Soaked At 5 Mm
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: CELL CYCLE Ligands: A1II4, EDO, EPE, PEG, GOL, ZN |
 |
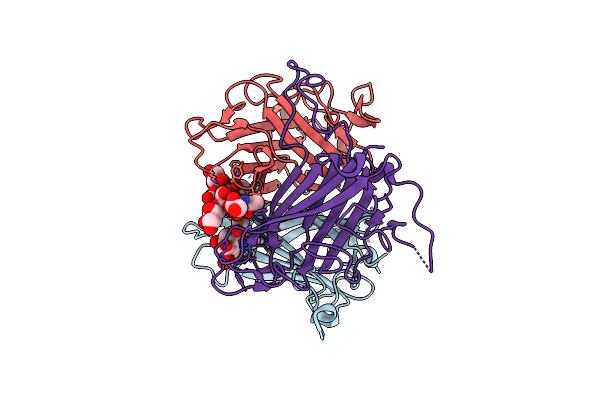
P53-Y220C Core Domain Covalently Bound To 3,5-Dichloro-6-Ethylpyrazine-2-Carbonitirle Soaked At 5 Mm
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: CELL CYCLE Ligands: A1II1, PEG, EDO, EPE, ZN, GOL, PGE |
 |
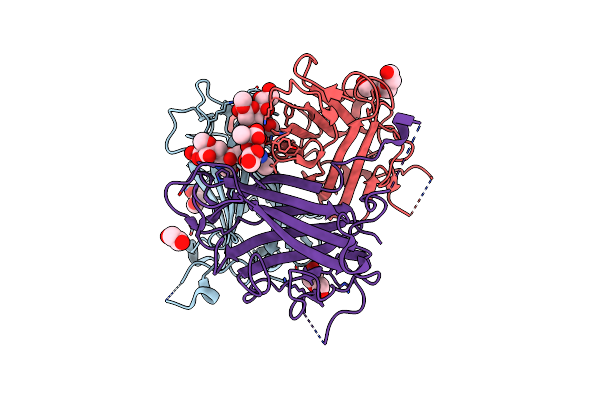
Human Adenovirus Type 36 Fiber Knob In Complex With 4-O-Acetyl-3'-Sialyllactose
Organism: Human adenovirus 36
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: PKM |
 |
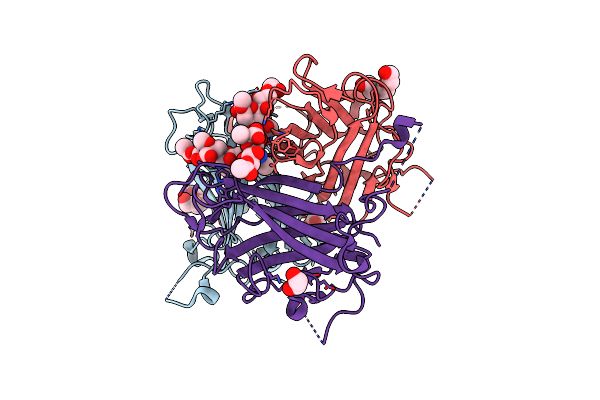
Human Adenovirus Type 36 Fiber Knob In Complex With 4-O,5-N-Diacetylneuraminic Acid
Organism: Human adenovirus 36
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: ANA, PEG, PGE, EDO |
 |
Human Adenovirus Type 36 Fiber Knob In Complex With 4,9-O,5-N-Triacetylneuraminic Acid
Organism: Human adenovirus 36
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: SIO, PGE, EDO |
 |
Human Adenovirus Type 37 Fiber Knob In Complex With 4-O,5-N-Diacetylneuraminic Acid
Organism: Human adenovirus d37
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: ANA, ZN, ACT, CL |
 |
P53-Y220C Core Domain Covalently Bound To 2,5,6-Trifluoropyridine-3-Carbonitrile Soaked At 5 Mm
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-11-27 Classification: CELL CYCLE Ligands: YLW, PEG, EDO, EPE, ZN, GOL |
 |
P53-Y220C Core Domain Covalently Bound To 2,5,6-Trifluoropyridine-3-Carbonitrile Soaked At 40 Mm
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-11-27 Classification: CELL CYCLE Ligands: YLW, GOL, EDO, EPE, ZN |
 |
P53-Y220C Core Domain Covalently Bound To 3-Amino-5-Chloropyrazine-2,6-Dicarbonitrile Soaked At 5 Mm
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-11-27 Classification: CELL CYCLE Ligands: YM0, EPE, PEG, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-09-25 Classification: TRANSFERASE Ligands: YOX, 1PE, PG4, EPE, PEG, GOL, EDO, SO4, PO4 |
 |
Organism: Streptomyces katrae
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Cytochrome P450 Gymb5 From Streptomyces Katrae In Complex With Cyy And Hypoxanthine
Organism: Streptomyces katrae
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: HEM, YTT, HPA, EDO |
 |
Organism: Streptomyces flavidovirens dsm 40150
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: HEM, GOL |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: CL, PO4, UPG, EDO, PG4, PGE |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: BJT, CL, BME, UDP, GOL |
 |
Jnk3 (Mitogen-Activated Protein Kinase 10) In Complex With Compound 23 Bearing A C(Sp3)F2Br Moiety
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: SWM, EDO, GOL, BME, PEG, C15, CL |

