Search Count: 20
 |
Acinetobacter Baumannii Tse15 Rhs Effector, Toxin Cleavage Mutant (D1369N, D1391N)
Organism: Acinetobacter baumannii ab307-0294
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: TOXIN |
 |
Organism: Acinetobacter baumannii ab307-0294
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: TOXIN |
 |
Non-Ribosomal Pcp-C Didomain (Amide Stabilised Leucine) Acceptor Bound State
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-21 Classification: BIOSYNTHETIC PROTEIN Ligands: YCK |
 |
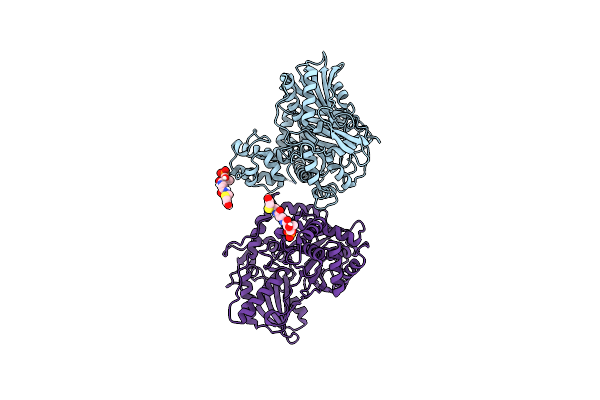
Non-Ribosomal Pcp-C Didomain (Ester Stabilised Leucine) Acceptor Bound State
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-21 Classification: BIOSYNTHETIC PROTEIN Ligands: YCT |
 |
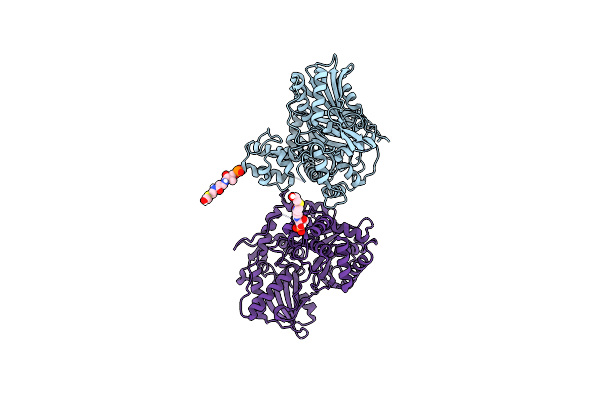
Non-Ribosomal Pcp-C Didomain (Thioether Stabilised Glycolic Acid) Acceptor Bound State
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2023-06-14 Classification: BIOSYNTHETIC PROTEIN Ligands: YJI |
 |
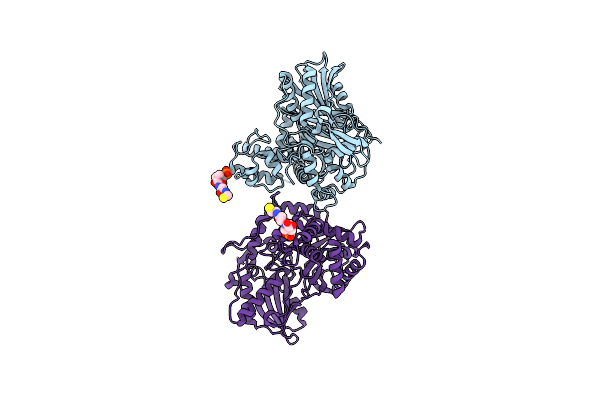
Non-Ribosomal Pcp-C Didomain R2577G (Thioether Stabilised Glycolic Acid) Acceptor Bound State
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-06-14 Classification: BIOSYNTHETIC PROTEIN Ligands: YJI |
 |
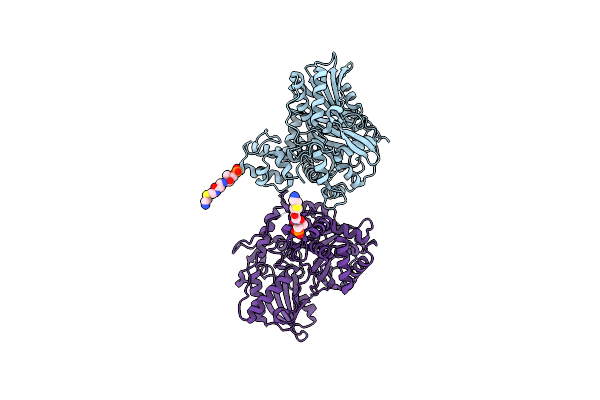
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-03-24 Classification: BIOSYNTHETIC PROTEIN Ligands: PNS |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-03-24 Classification: BIOSYNTHETIC PROTEIN Ligands: XAG |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-03-24 Classification: BIOSYNTHETIC PROTEIN Ligands: PNS |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-03-24 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2020-05-27 Classification: TRANSFERASE Ligands: GOL, ACT, CL |
 |
Crystal Structure Of Beta-Ketoadipyl-Coa Thiolase Mutant (H356A) In Complex With Coa
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2020-05-27 Classification: TRANSFERASE Ligands: COA, GOL, CL |
 |
Crystal Structure Of Beta-Ketoadipyl-Coa Thiolase Mutant (H356A) In Complex Hexanoyl Coenzyme A
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-05-27 Classification: TRANSFERASE Ligands: COA, O8Y, GOL |
 |
Crystal Structure Of Beta-Ketoadipyl-Coa Thiolase Mutant (C90S-H356A) In Complex Octanoyl Coenzyme A
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2020-05-27 Classification: TRANSFERASE Ligands: COA, OYA, GOL, CL, K |
 |
Double Cache (Dcache) Sensing Domain Of Tlpc Chemoreceptor From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2017-11-08 Classification: SIGNALING PROTEIN Ligands: LAC, GOL |
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-03-20 Classification: TOXIN Ligands: MG, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2012-03-28 Classification: HYDROLASE Ligands: CL, K, BCT, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:5.20 Å Release Date: 2012-03-28 Classification: HYDROLASE |
 |
Crystal Structure Of The Basic Protease Bprv From The Ovine Footrot Pathogen, Dichelobacter Nodosus
Organism: Dichelobacter nodosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of The Basic Protease Bprb From The Ovine Footrot Pathogen, Dichelobacter Nodosus
Organism: Dichelobacter nodosus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: CA, GOL, CL |

