Search Count: 27
 |
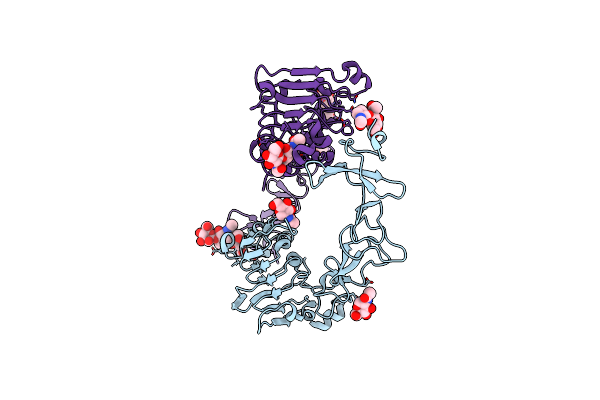
Crystal Structure Of Nanobody/Vhh Domain Of 34E5 In Complex With The Extracellular Region Of The Epidermal Growth Factor Variant Iii (Egfrviii)
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2024-06-19 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: NAG |
 |
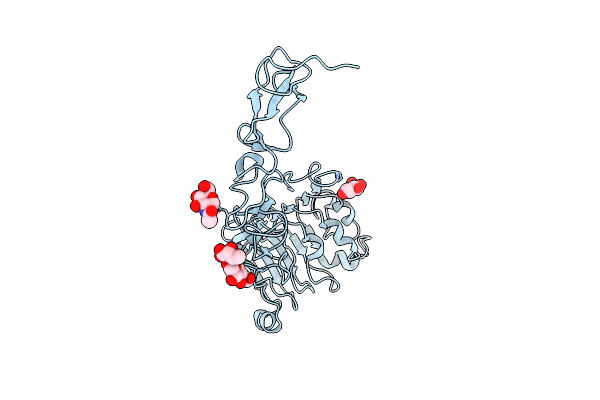
Crystal Structure The Extracellular Region Of The Epidermal Growth Factor Receptor Variant Iii (Egfrviii) At Ph 5.0
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: NAG |
 |
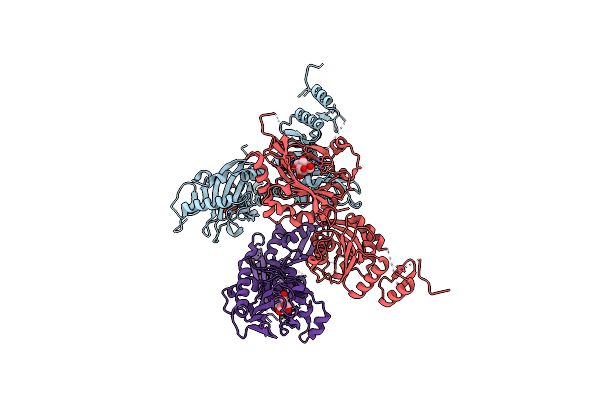
Crystal Structure The Extracellular Region Of The Epidermal Growth Factor Receptor Variant Iii (Egfrviii) At Ph 7.0
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: NAG |
 |
Organism: Micromonospora carbonacea
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2018-12-26 Classification: TRANSFERASE, oxidoreductase Ligands: NI, BTB |
 |
Escherichia Coli Quinol:Fumarate Reductase Crystallized Without Dicarboxylate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2017-12-06 Classification: ELECTRON TRANSPORT Ligands: FAD, PO4, FES, F3S, SF4 |
 |
Organism: Micromonospora carbonacea
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2017-09-06 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of The N-Terminal Domain Of Evdmo1 In The Presence Of Sah And D-Fucose
Organism: Micromonospora carbonacea
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-09-06 Classification: TRANSFERASE Ligands: SAH |
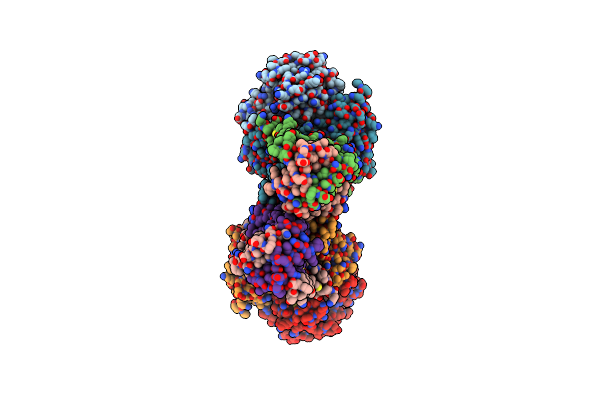 |
Organism: Escherichia coli (strain k12), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:4.22 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, FES, F3S, SF4 |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, GOL |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, TRS, ACT, HSX |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, GOL, 1X4 |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, GOL |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, 1X4, GOL, ACT |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, GOL, ACT |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, TRS, GOL |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, 1X4 |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-31 Classification: ISOMERASE Ligands: MN, GOL, HSX |
 |
Structure Of The Chey-Bef3 Complex With Substitutions At 59 And 89: N59D And E89Q
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-05-09 Classification: SIGNALING PROTEIN Ligands: MN, BEF, SO4, GOL |
 |
Structure Of The Chey-Mn2+ Complex With Substitutions At 59 And 89: N59D E89Q
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2012-05-09 Classification: SIGNALING PROTEIN Ligands: MN |
 |
Structure Of The Chey-Bef3 Complex With Substitutions At 59 And 89: N59D And E89R
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-05-09 Classification: SIGNALING PROTEIN Ligands: BEF, MN, SO4, GOL |

