Search Count: 20
 |
Pressure Wave-Exposed Human Hemoglobin: Probe Only Data (3500 Indexed Images)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2021-02-17 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
 |
Pressure Wave-Exposed Human Hemoglobin: Probe Only Data (5500 Indexed Images)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2021-02-17 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
 |
Pressure Wave-Exposed Human Hemoglobin: Pump/Probe Data (3500 Indexed Images)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2021-02-17 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
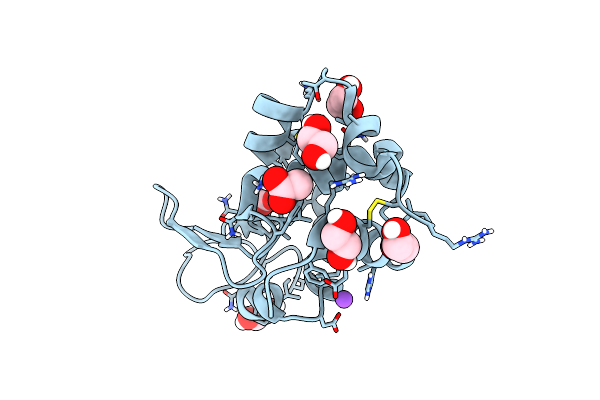 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2018-10-10 Classification: HYDROLASE Ligands: CL, NA, ACT, EDO |
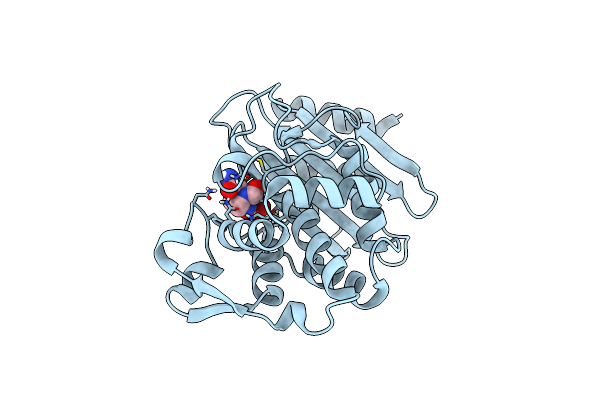 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2018-10-10 Classification: ANTIBIOTIC Ligands: NXL |
 |
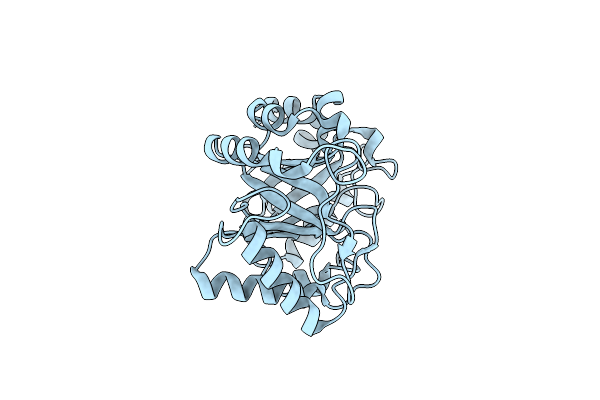
Concanavalin A Structure Determined With Data From The Euxfel, The First Mhz Free Electron Laser
Organism: Canavalia cathartica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-05 Classification: SUGAR BINDING PROTEIN Ligands: MG, CA |
 |
Concanavalin B Structure Determined With Data From The Euxfel, The First Mhz Free Electron Laser
Organism: Canavalia ensiformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-05 Classification: SUGAR BINDING PROTEIN |
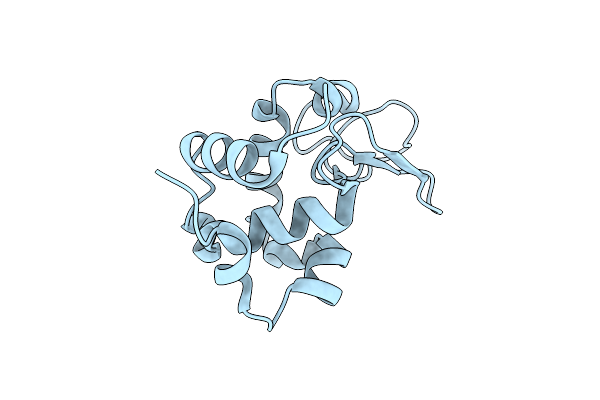 |
Hen Egg-White Lysozyme Structure Determined With Data From The Euxfel, The First Mhz Free Electron Laser, 7.47 Kev Photon Energy
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-09-05 Classification: HYDROLASE |
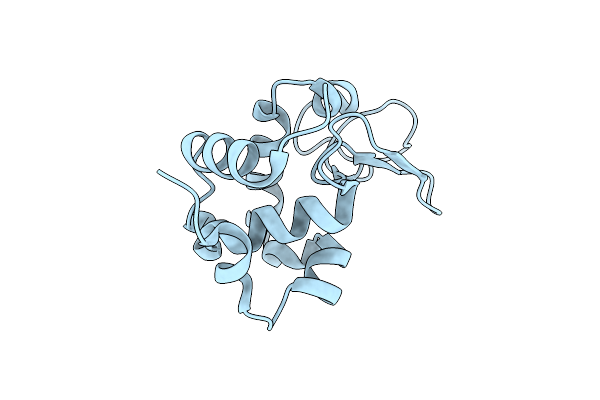 |
Hen Egg-White Lysozyme Structure Determined With Data From The Euxfel, 9.22 Kev Photon Energy
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-05 Classification: HYDROLASE |
 |
Structure Of Pas-Gaf Fragment Of Deinococcus Phytochrome By Serial Femtosecond Crystallography
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-02-22 Classification: TRANSFERASE Ligands: LBV, NI, CL, EDO |
 |
Structure Of The Photosensory Module Of Deinococcus Phytochrome By Serial Femtosecond X-Ray Crystallography
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-02-22 Classification: TRANSFERASE Ligands: LBV |
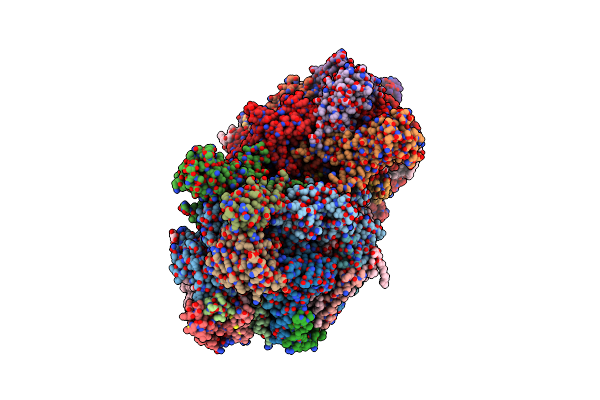 |
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-11-23 Classification: ELECTRON TRANSPORT Ligands: OEX, FE2, SQD, CL, CLA, PHO, BCR, PL9, LMG, UNL, LHG, BCT, DGD, HEM, HEC |
 |
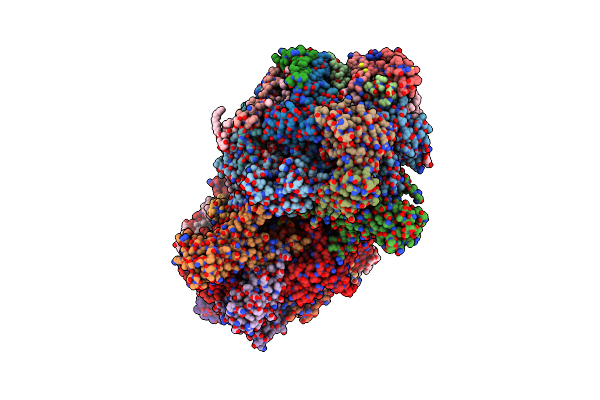
Nh3-Bound Rt Xfel Structure Of Photosystem Ii 500 Ms After The 2Nd Illumination (2F) At 2.8 A Resolution
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-23 Classification: ELECTRON TRANSPORT Ligands: OEX, FE2, LMG, CL, CLA, PHO, BCR, PL9, SQD, UNL, BCT, LHG, DGD, HEM, HEC |
 |
Room Temperature Xfel Structure Of The Native, Doubly-Illuminated Photosystem Ii Complex
Organism: Thermosynechococcus elongatus (strain bp-1), Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-11-23 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, CL, BCT, CLA, PHO, BCR, PL9, SQD, LHG, UNL, LMG, DGD, HEM, HEC |
 |
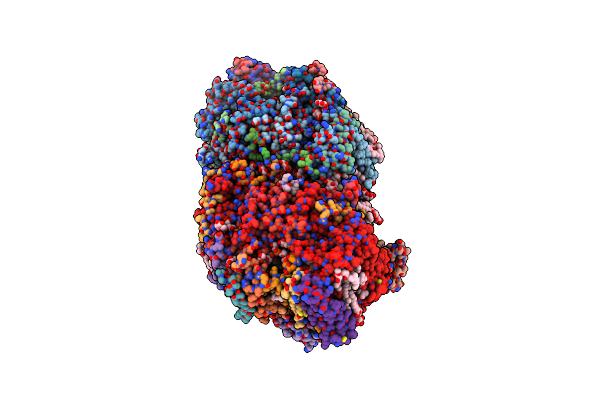
Ambient-Temperature Crystal Structure Of 30S Ribosomal Subunit From Thermus Thermophilus In Complex With Paromomycin
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-11-18 Classification: RIBOSOME Ligands: PAR, MG, ZN |
 |
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:4.90 Å Release Date: 2014-07-09 Classification: ELECTRON TRANSPORT,PHOTOSYNTHESIS Ligands: FE2, CLA, PL9, BCR, DGD, LHG, LMG, CL, OEX, SQD, LMT, PHO, BCT, HEM, CA |
 |
Rt Xfel Structure Of Photosystem Ii 500 Ms After The Third Illumination At 4.6 A Resolution
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2014-07-09 Classification: ELECTRON TRANSPORT,PHOTOSYNTHESIS Ligands: FE2, CLA, PL9, BCR, DGD, LHG, LMG, OEX, SQD, LMT, PHO, CL, BCT, HEM, CA |
 |
Rt Xfel Structure Of Photosystem Ii 500 Ms After The 2Nd Illumination (2F) At 4.5 A Resolution
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2014-07-09 Classification: ELECTRON TRANSPORT,PHOTOSYNTHESIS Ligands: FE2, CLA, PL9, BCR, DGD, LHG, LMG, CL, OEX, SQD, LMT, PHO, BCT, HEM, CA |
 |
Rt Xfel Structure Of Photosystem Ii 250 Microsec After The Third Illumination At 5.2 A Resolution
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:5.20 Å Release Date: 2014-07-09 Classification: ELECTRON TRANSPORT,PHOTOSYNTHESIS Ligands: FE2, CLA, PHO, PL9, BCR, DGD, LHG, CL, OEX, SQD, LMG, LMT, BCT, HEM, CA |
 |
1.8 A Resolution Room Temperature Structure Of Thermolysin Recorded Using An Xfel
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: ZN, CA |

