Search Count: 461
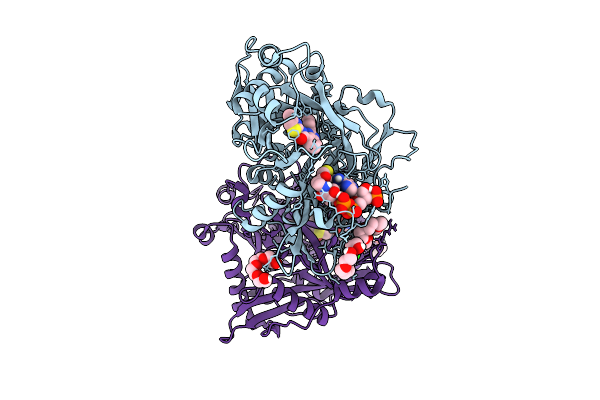 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20045
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHP, CL, PG4, 1PE, PGE |
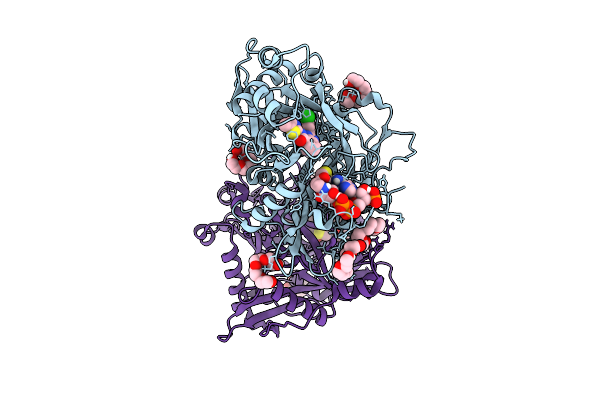 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20057
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHQ, CL, 1PE, PG4 |
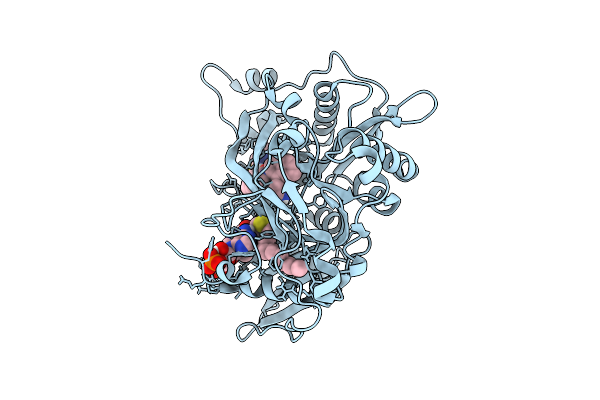 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20084
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHR, CL |
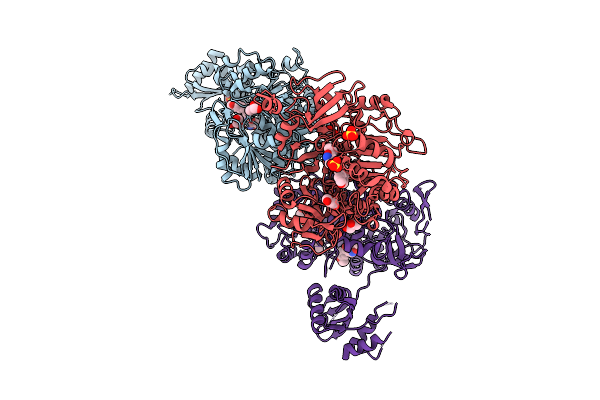 |
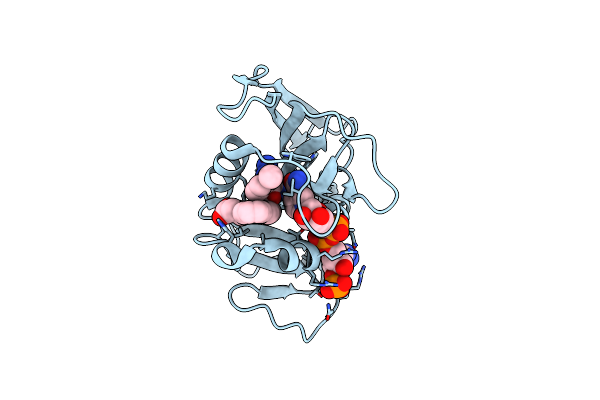
Crystal Structure Of Acetyl-Coa Synthetase From Cryptococcus Neoformans H99 In Complex With Inhibitor Hgn-1310 (Dd3-027)
Organism: Cryptococcus neoformans var. grubii
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LIGASE Ligands: GOL, SO4, CL, A1AV1 |
 |
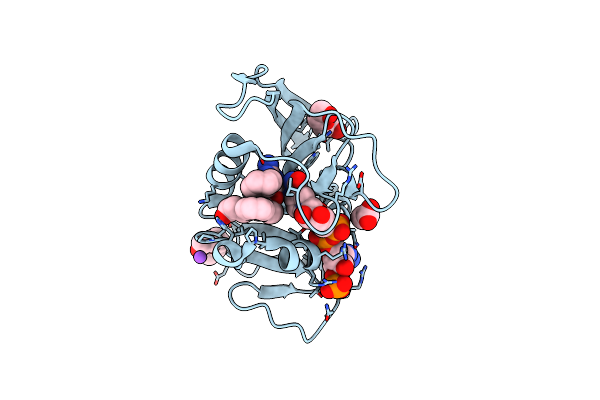
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam881
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: DQI, NAP |
 |
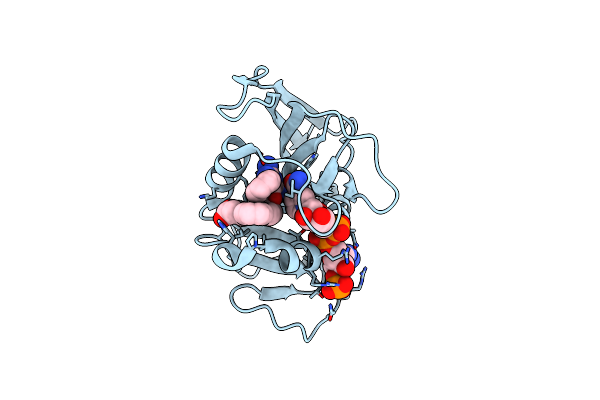
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam907
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE/INHIBITOR |
 |
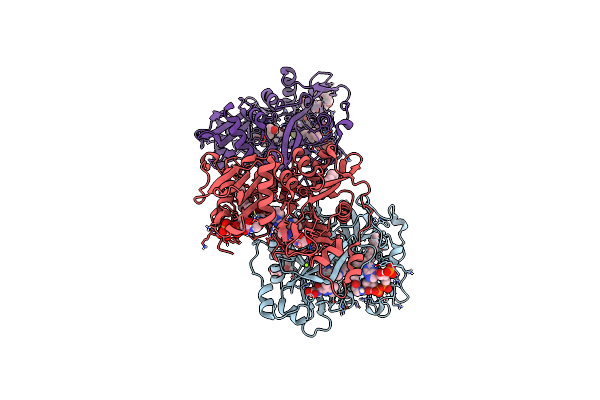
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam758
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, HIJ |
 |
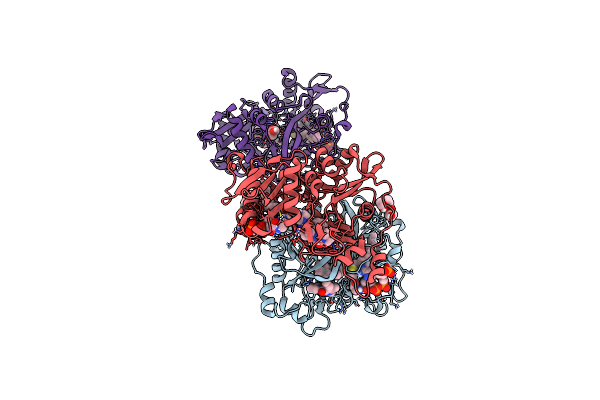
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 9C
Organism: Plasmodium vivax sal-1
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-17 Classification: TRANSFERASE Ligands: MYA, A1AB7, CL, GOL, 1PE, MG, P33 |
 |
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 10B
Organism: Plasmodium vivax sal-1
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-07-17 Classification: TRANSFERASE Ligands: MYA, A1AB8, CL, GOL, 1PE, P6G |
 |
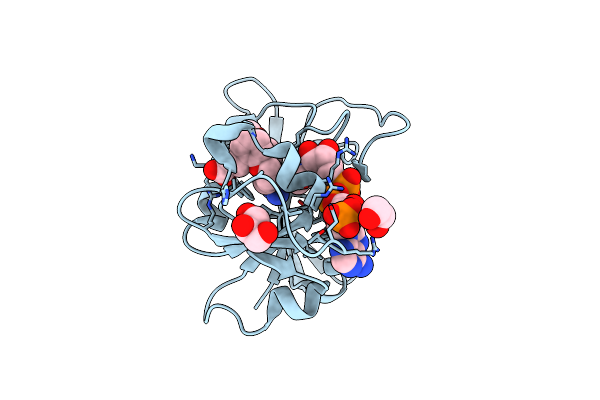
Crystal Structure Of Glutamyl-Trna Synthetase Glurs From Pseudomonas Aeruginosa (Zinc Bound)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-27 Classification: LIGASE Ligands: FLC, SO4, ZN, 2PE, GOL |
 |
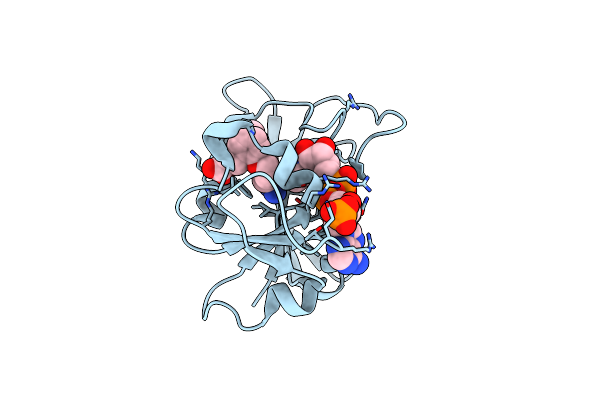
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam738
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XJB, GOL, EDO |
 |
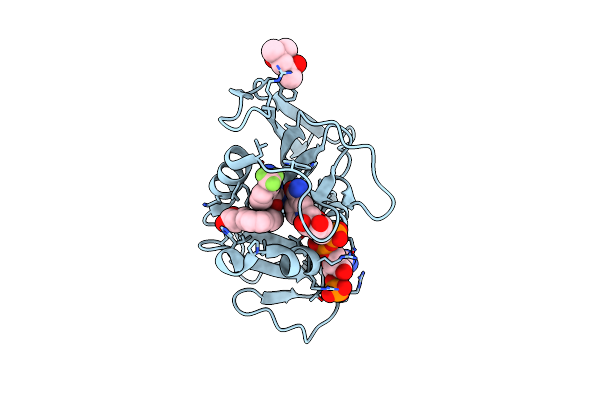
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam777
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XJN |
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam845
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: XJT, MPD, NAP, BR |
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam846
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XJZ, IMD |
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam787
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XK8 |
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam851
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XKI |
 |
Crystal Structure Of Acetyl-Coa Synthetase From Cryptococcus Neoformans H99 In Complex With An Ethylsulfamide Amp Inhibitor
Organism: Cryptococcus neoformans var. grubii
Method: X-RAY DIFFRACTION Release Date: 2023-12-13 Classification: LIGASE Ligands: YDO, CL |
 |
Crystal Structure Of Acetyl-Coa Synthetase 2 In Complex With Amp And Coa From Candida Albicans
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-06 Classification: LIGASE Ligands: AMP, COA, ACY, GOL |
 |
Crystal Structure Of Plasmodium Vivax Glycylpeptide N-Tetradecanoyltransferase (N-Myristoyltransferase, Nmt) Bound To Myristoyl-Coa And Inhibitor 12B
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-10-18 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOQ, GOL, SO4, CL, ACT, PEG, 1PE, PG4, PGE |
 |
Crystal Structure Of Glycine--Trna Ligase From Mycobacterium Tuberculosis (G5A Bound)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2023-09-13 Classification: TRANSFERASE Ligands: G5A, IMD, MES, EDO, MG, CL |

