Search Count: 88
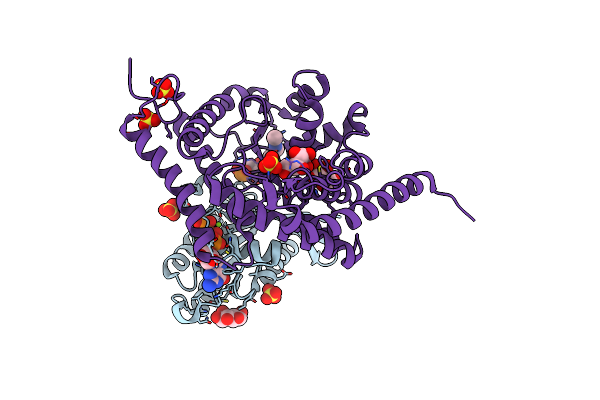 |
Organism: Homo sapiens, Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2020-12-23 Classification: CELL INVASION Ligands: MG, GNP, GLC, LJN, SO4 |
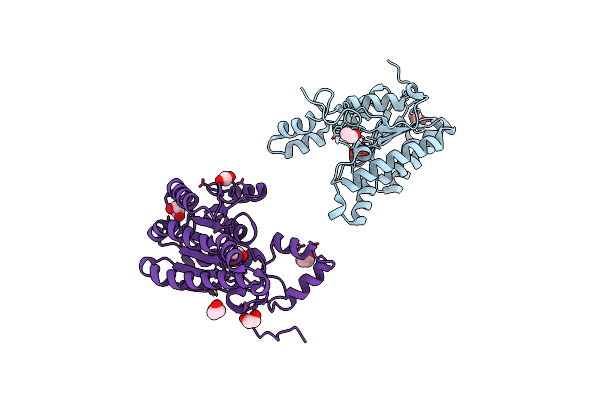 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: EDO |
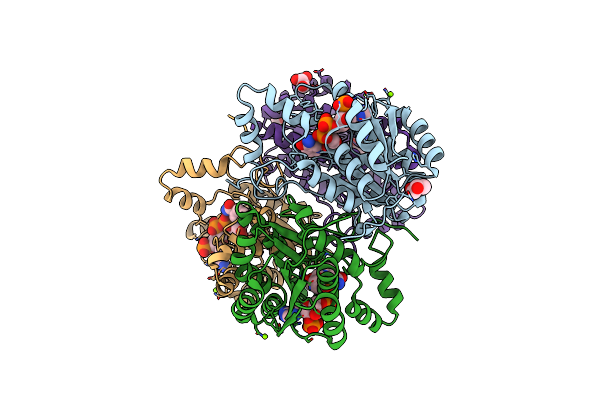 |
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: NAP, GOL, MG |
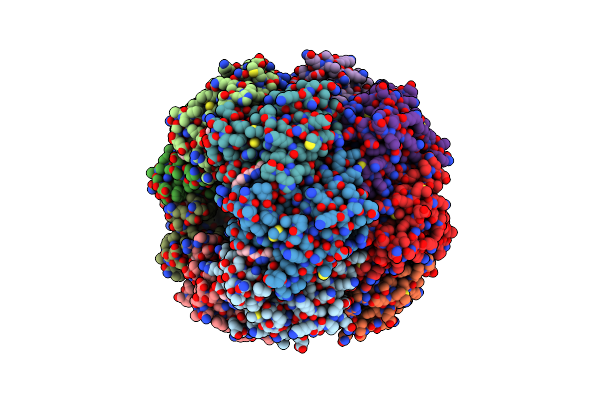 |
Crystal Structure Of The Hclpp Y118A Mutant With An Activating Small Molecule
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: FJT, EDO |
 |
Organism: Staphylococcus aureus (strain mu50 / atcc 700699)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, GOL |
 |
Structure Of The Alanine Racemase From Staphylococcus Aureus In Complex With A Pyridoxal 5' Phosphate-Derivative
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: EOW, NA, MPD, ACT, CL, MRD, GOL |
 |
Structure Of The Alanine Racemase From Staphylococcus Aureus In Complex With An Pyridoxal-6- Phosphate Derivative
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: EM2, SO4, NA, CL |
 |
Organism: Corallococcus coralloides
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-29 Classification: HYDROLASE Ligands: MPD |
 |
Crystal Structure Of Corb Derivatized With S-(2-Acetamidoethyl) 4-Methyl-3-Oxohexanethioate
Organism: Corallococcus coralloides
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2016-03-30 Classification: HYDROLASE Ligands: MPD, NA |
 |
Organism: Corallococcus coralloides
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2016-03-30 Classification: HYDROLASE |
 |
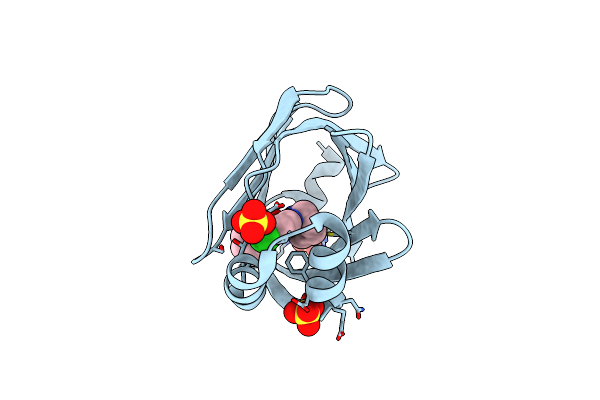
Human Fatty Acid Binding Protein 4 In Complex With 6-Chloro-2-Methyl-4-Phenyl-Quinoline-3-Carboxylic Acid At 1.18A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2016-03-09 Classification: LIPID BINDING PROTEIN Ligands: 5M8 |
 |
Human Fabp4 In Complex With 6-Chloro-4-Phenyl-2-Piperidin-1-Yl-Quinoline-3-Carboxylic Acid At 1.29A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2016-03-09 Classification: LIPID BINDING PROTEIN Ligands: 5M7, SO4 |
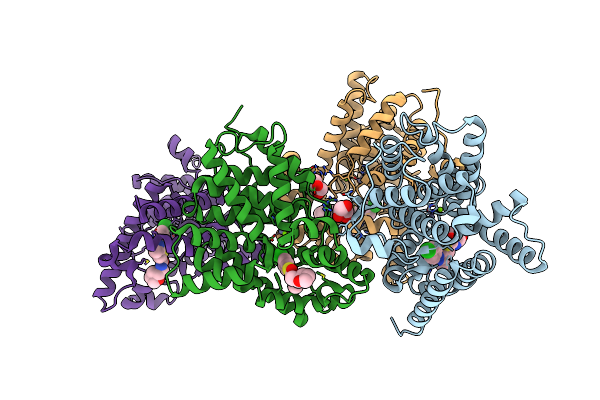 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With C13C(Cc(S1)C(Ncc2Occc2)=O)C(Nn3C4Ccc(Cc4)Cl)C, Micromolar Ic50=0.217
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, GOL, 5M6 |
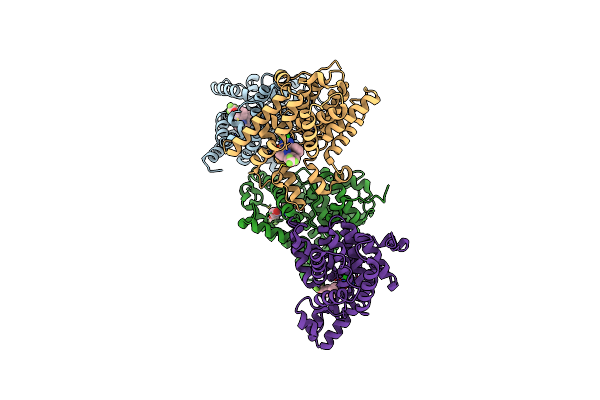 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With C1(C(Nc([Nh]1)Cl)C2Ccccc2)C4=Nn(C3Cccc(C3)Oc(F)(F)F)C=Cc4=O, Micromolar Ic50=0.029618
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, 5MG |
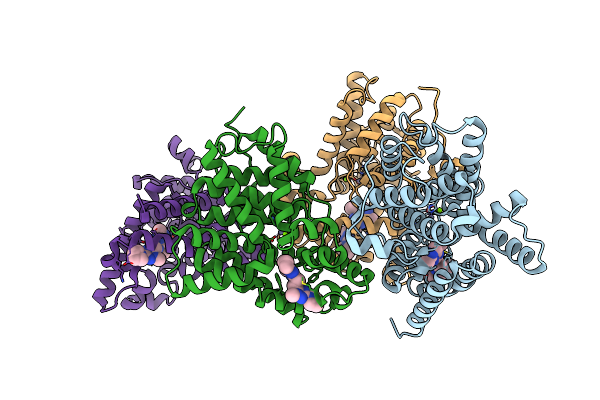 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With N4C(C)N1C(Nc(N1)Ccc2Nc(Nn2C)N3Cccc3)C(C4)Cc, Micromolar Ic50=0.0037753
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, 5MF |
 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With C2(C(N1Nc(Nc1C(C2)C)Ccc3Nc(Nn3C)N4Cccc4)C)Cl, Micromolar Ic50=0.000279
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, 5M9 |
 |
Crystal Structure Of Human Phosphodiesterase 10 In Complex With C2(=Nn(C1Cccc(C1)Oc(F)(F)F)C=Cc2=O)C3Ccnn3C4Ccccc4, Micromolar Ic50=0.019462
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-03-09 Classification: HYDROLASE Ligands: ZN, MG, 67A |
 |
Crystal Structure Of Bace-1 In Complex With {(1R,2R)-2-[(R)-2-Amino-4-(4-Difluoromethoxy-Phenyl)-4,5-Dihydro-Oxazol-4-Yl]-Cyclopropyl}-(5-Chloro-Pyridin-3-Yl)-Methanone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-02-24 Classification: Hydrolase/Inhibitor Ligands: 5T5, NA, DMS |
 |
Crystal Structure Of Bace-1 In Complex With (4S)-4-[3-(5-Chloro-3-Pyridyl)Phenyl]-4-[4-(Difluoromethoxy)-3-Methyl-Phenyl]-5H-Oxazol-2-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-02-24 Classification: Hydrolase/Inhibitor Ligands: 5T6, DMS, NA |
 |
Crystal Structure Of Bace-1 In Complex With 5-[3-[(3-Chloro-8-Quinolyl)Amino]Phenyl]-5-Methyl-2,6-Dihydro-1,4-Oxazin-3-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-02-24 Classification: HYDROLASE |

