Search Count: 41
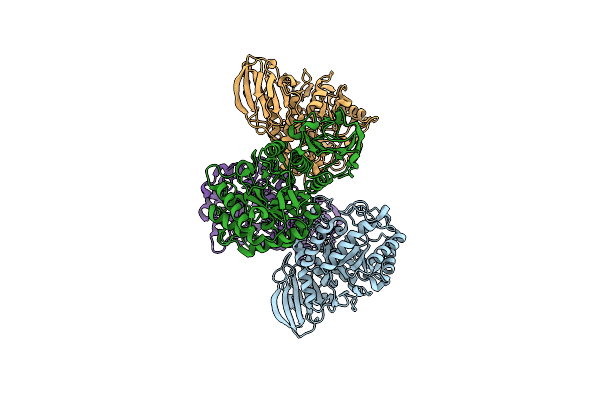 |
Crystal Structure Of A Gh140 Apiosidase Derived From A Lignocellulolytic Enriched Mangrove Metagenome.
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-07 Classification: HYDROLASE |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-15 Classification: HYDROLASE Ligands: BR |
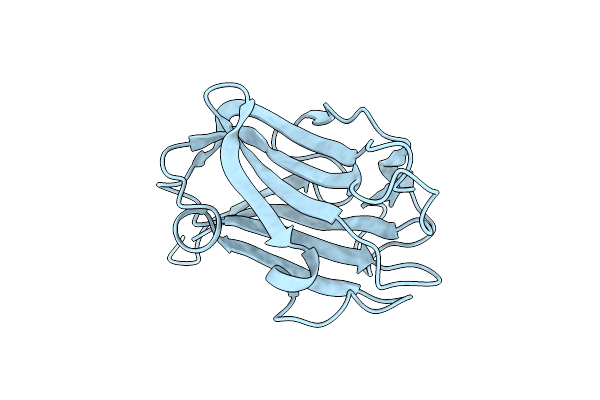 |
The Crystal Structure Of Family 8 Carbohydrate-Binding Module From Dictyostelium Discoideum Complexed With Iodine Atoms
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: IOD |
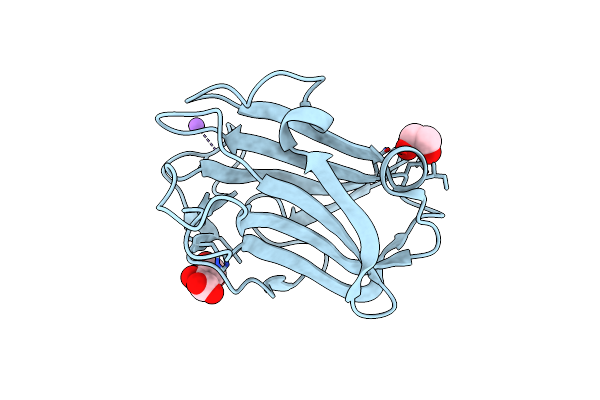 |
The Crystal Structure Of Family 8 Carbohydrate-Binding Module From Dictyostelium Discoideum
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: EDO, NA, LMR |
 |
Crystal Structure Of Sclam, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA, GOL |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With 1,3-Beta-D-Cellotriosyl-Glucose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With 1,3-Beta-D-Cellobiosyl-Cellobiose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Cellotriose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose And A Cellobiose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Cellohexaose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Laminarihexaose, Presenting A Laminaribiose And A Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: PO4, GLC, CA |
 |
Organism: Thermotoga petrophila rku-1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: MPD, EDO, ACT, CO3, GOL, PEG, TRS, CL, NA |
 |
Structure Of An Enoyl-Coa Hydratase/Aldolase Isolated From A Lignin-Degrading Consortium
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-08 Classification: LYASE Ligands: COA, B3P |
 |
Structural And Functional Characterisation Of A Bacterial Laccase-Like Multi-Copper Oxidase Cueo From Lignin-Degrading Bacterium Ochrobactrum Sp. With Oxidase Activity Towards Lignin Model Compounds And Lignosulfonate
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-04-11 Classification: OXIDOREDUCTASE Ligands: CU, EDO |
 |
Structure Of An Alpha-L-Arabinofuranosidase (Gh62) From Aspergillus Nidulans
Organism: Emericella nidulans (strain fgsc a4 / atcc 38163 / cbs 112.46 / nrrl 194 / m139)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-09-27 Classification: HYDROLASE Ligands: CA |
 |
Organism: Coptotermes gestroi
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2017-01-18 Classification: OXIDOREDUCTASE Ligands: NAP |
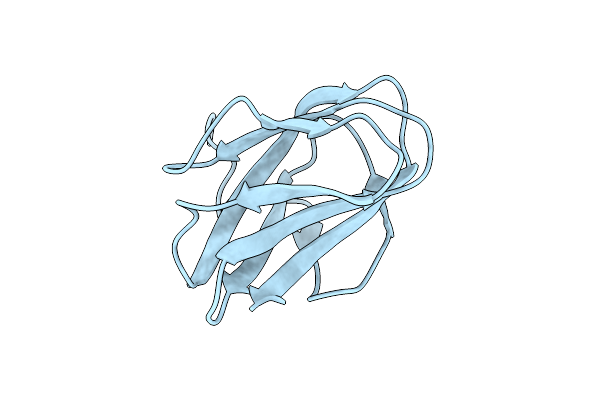 |
Structure Of Cbm_E1, A Novel Carbohydrate-Binding Module Found By Sugar Cane Soil Metagenome
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-09-21 Classification: SUGAR BINDING PROTEIN |
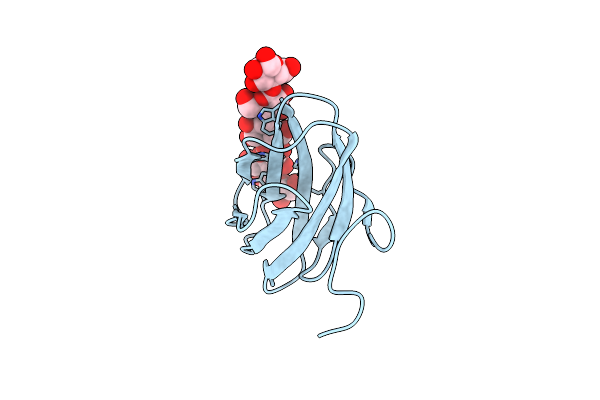 |
Structure Of Cbm_E1, A Novel Carbohydrate-Binding Module Found By Sugar Cane Soil Metagenome, Complexed With Cellopentaose
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-09-21 Classification: SUGAR BINDING PROTEIN |
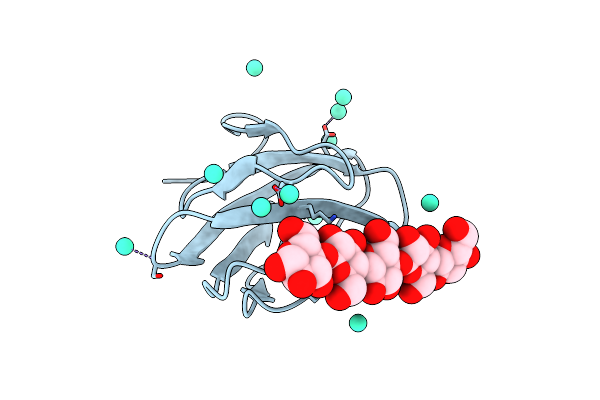 |
Structure Of Cbm_E1, A Novel Carbohydrate-Binding Module Found By Sugar Cane Soil Metagenome, Complexed With Cellopentaose And Gadolinium Ion
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-21 Classification: SUGAR BINDING PROTEIN Ligands: GD |
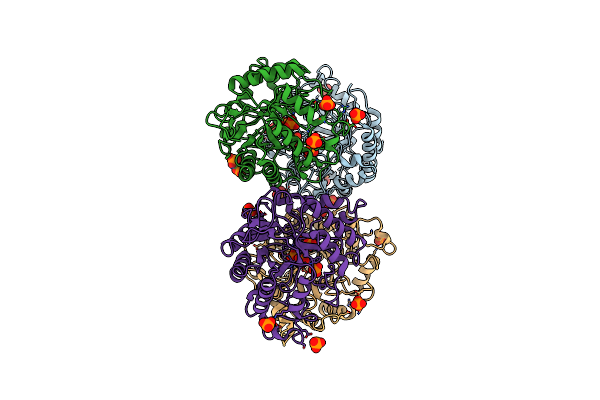 |
Catalytic Domain Of The Major Endoglucanase From Xanthomonas Campestris Pv. Campestris
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-10-14 Classification: HYDROLASE Ligands: 2HP |
 |
Crystal Structure Of Endo-1,5-Alpha-L-Arabinanase From Thermotoga Petrophila Rku-1
Organism: Thermotoga petrophila
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-02-05 Classification: HYDROLASE Ligands: PEG, CA |

