Search Count: 15
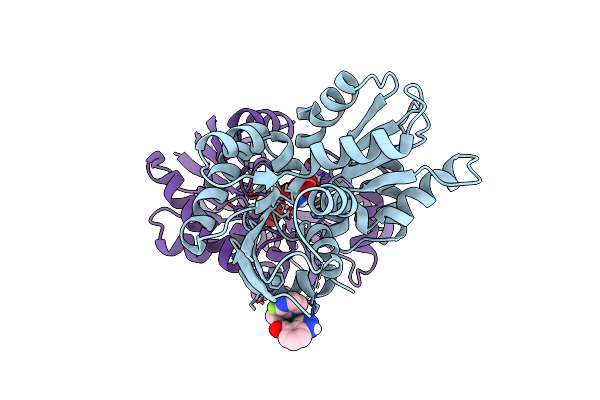 |
Crystal Structure Of Helicobacter Pylori Glutamate Racemase Bound To D-Glutamate And A Crystallographic Artifact
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-09 Classification: ISOMERASE Ligands: DGL, WAW, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: RECOMBINATION |
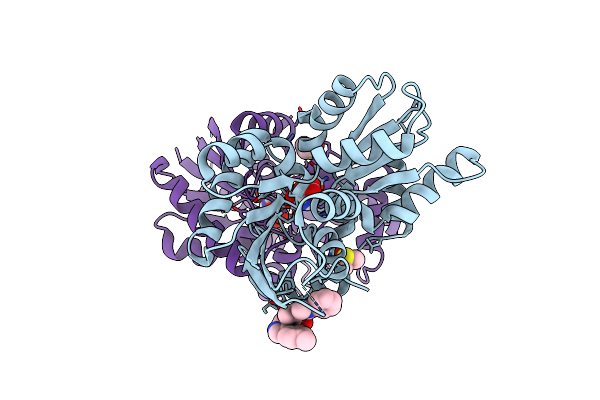 |
Crystal Structure Of Helicobacter Pylori Glutamate Racemase Bound To D-Glutamate And A Crystallographic Artifact
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-07-03 Classification: ISOMERASE Ligands: ZMX, DGL, DMS |
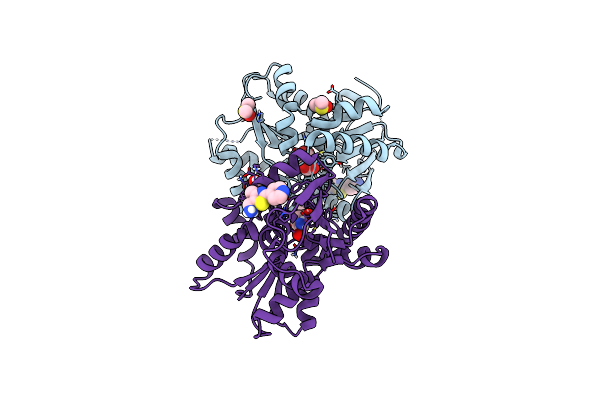 |
Crystal Structure Of Glutamate Racemase From Helicobacter Pylori In Complex With A Fragment
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: GOL, DMS, WFI, DGL, CL |
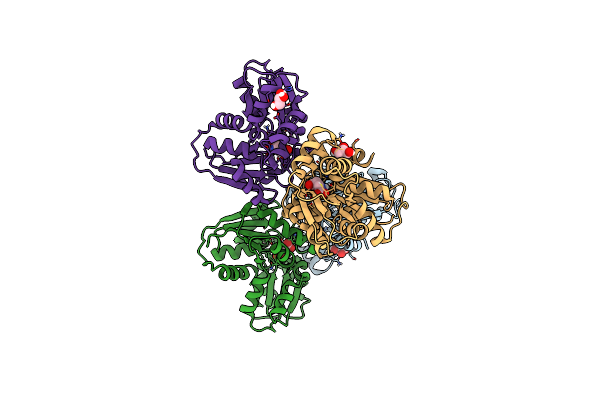 |
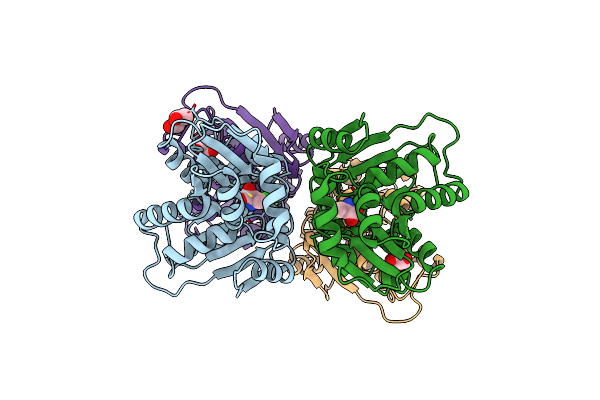
Crystal Structure Of Glutamate Racemase From Helicobacter Pylori In Complex With D-Glutamate
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-04-05 Classification: ISOMERASE Ligands: DGL, GOL |
 |
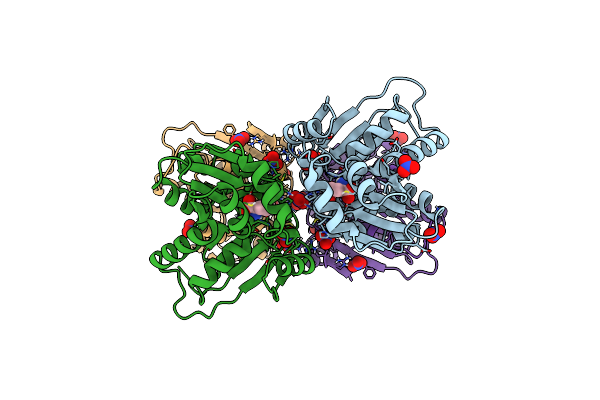
Crystal Structure Of Glutamate Racemase From Thermus Thermophilus In Complex With Beta-Chloro-D-Alanine
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-06-05 Classification: ISOMERASE Ligands: C2N, GOL |
 |
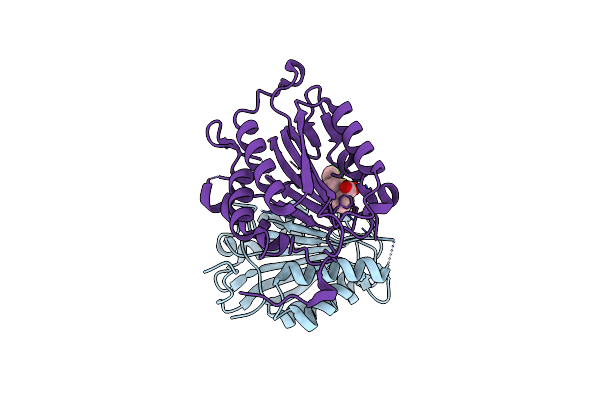
Crystal Structure Of Glutamate Racemase From Thermus Thermophilus In Complex With D-Glutamate
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2018-06-06 Classification: ISOMERASE Ligands: DGL, NO3, CL |
 |
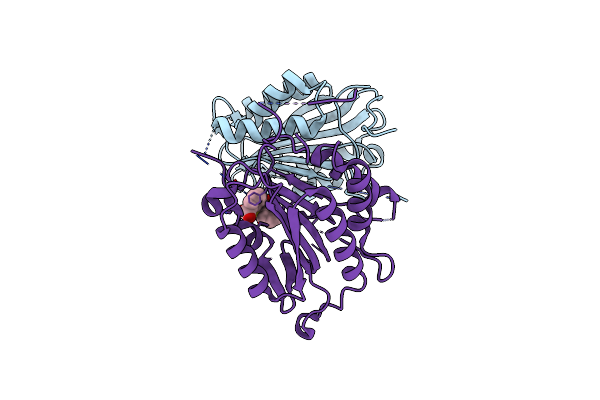
Crystal Structure Of Human Caspase-7 Soaked With Allosteric Inhibitor 2-{[2-(4-Chlorophenyl)-2-Oxoethyl]Sulfanyl}Benzoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-03-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 8YJ |
 |
Crystal Structure Of Human Caspase-7 Soaked With Allosteric Inhibitor 2-[(2-Acetylphenyl)Sulfanyl]Benzoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-10-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 8YM |
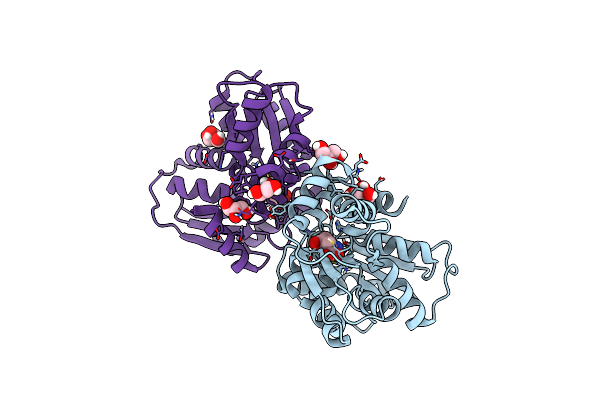 |
Crystal Structure Of Alternate Isoform Of Glutamate Racemase From Helicobacter Pylori Bound To D-Glutamate
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-06-21 Classification: ISOMERASE Ligands: DGL, GOL |
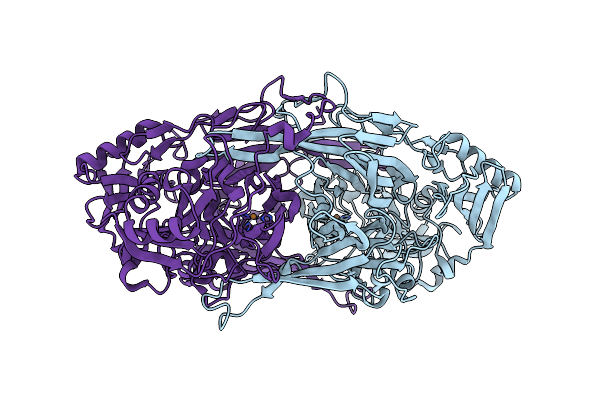 |
Crystal Structure Of D298K Copper Amine Oxidase From Arthrobacter Globiformis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2008-04-08 Classification: OXIDOREDUCTASE Ligands: CU |
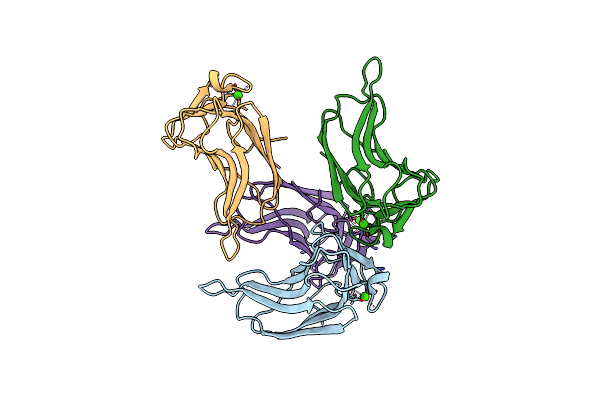 |
Organism: Thermoanaerobacterium polysaccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-03-25 Classification: HYDROLASE Ligands: CA |
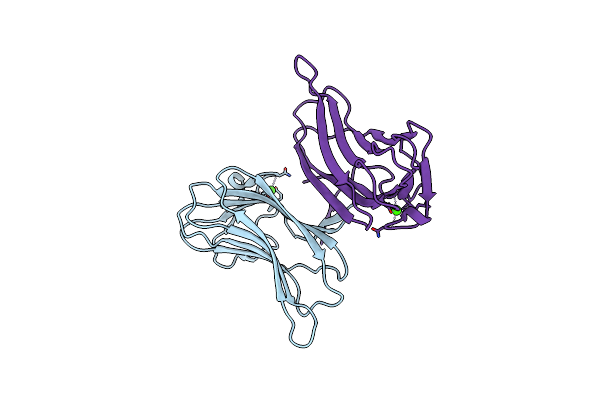 |
Organism: Thermoanaerobacterium polysaccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-03-04 Classification: HYDROLASE Ligands: CA |
 |
Organism: Thermoanaerobacterium polysaccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2008-03-04 Classification: HYDROLASE Ligands: CA |
 |
Organism: Thermoanaerobacterium polysaccharolyticum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-03-04 Classification: HYDROLASE Ligands: CA |

