Search Count: 499
All
Selected
 |
Organism: Novosphingopyxis baekryungensis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: CELL INVASION Ligands: MG |
 |
Organism: Bacteriophage sp., Novosphingopyxis baekryungensis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: CELL INVASION |
 |
Crystal Structure Of A (R)-Selective Transaminase (Rta) From Sphingopyxis Sp.
Organism: Sphingopyxis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-10-22 Classification: TRANSFERASE |
 |
Organism: Novosphingopyxis baekryungensis dsm 16222
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
 |
Organism: Novosphingopyxis baekryungensis dsm 16222
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG |
 |
Organism: Novosphingopyxis baekryungensis dsm 16222
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG |
 |
Organism: Novosphingopyxis baekryungensis dsm 16222
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG |
 |
Crystal Structure Of Fmn And Nadph-Dependent Nitroreductase Nfnb From Sphigopyxis Sp. Strain Hmh
Organism: Sphingopyxis sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-09-08 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Crystal Structure Of Fmn And Nadph-Dependent Nitroreductase Nfnb Mutant Y88A Derived From Sphigopyxis Sp. Strain Hmh
Organism: Sphingopyxis sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-09-08 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Crystal Structure Of Fmn And Nadph-Dependent Nitroreductase Nfnb Mutant Y88F Derived From Sphigopyxis Sp. Strain Hmh
Organism: Sphingopyxis sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-09-08 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Structure Of Plp Internal Aldimine Form Of Sphingopyxis Sp. Mta144 Fumi Protein
Organism: Sphingopyxis macrogoltabida
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-08-28 Classification: TRANSFERASE Ligands: PLP, MG |
 |
Structure Of Plp Internal Aldimine Form Of Sphingopyxis Sp. Mta144 Fumi Protein
Organism: Sphingopyxis macrogoltabida
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-08-28 Classification: TRANSFERASE Ligands: PLP, PMP, MG |
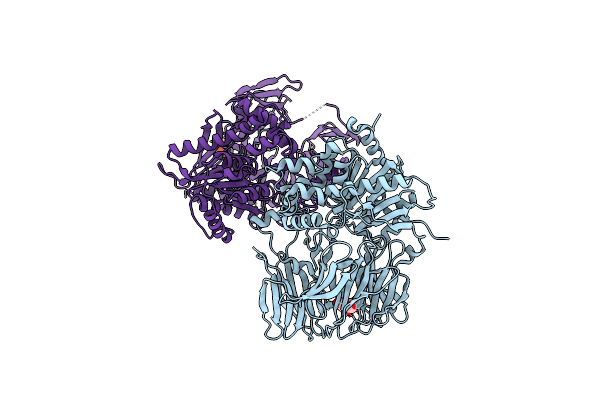 |
Crystal Structure Of The Sphingopyxin I Lasso Peptide Isopeptidase Spi-Isop (Semet-Derived)
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-09-28 Classification: HYDROLASE Ligands: BGC |
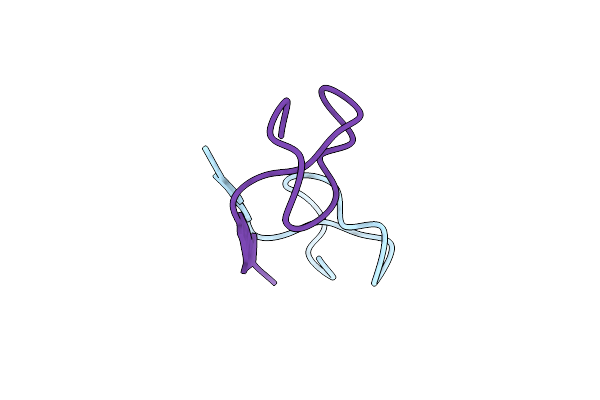 |
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2016-09-14 Classification: UNKNOWN FUNCTION |
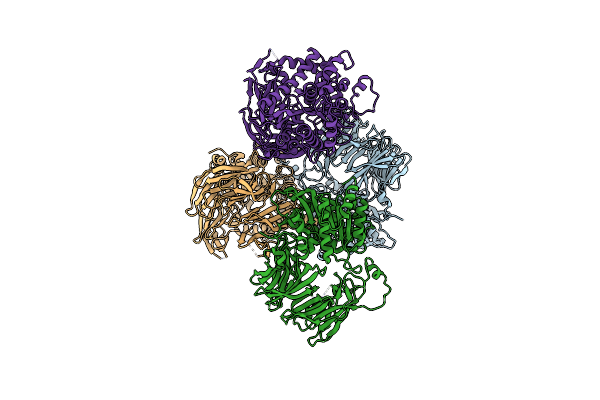 |
Crystal Structure Of The Sphingopyxin I Lasso Peptide Isopeptidase Spi-Isop (Native)
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-09-14 Classification: HYDROLASE |
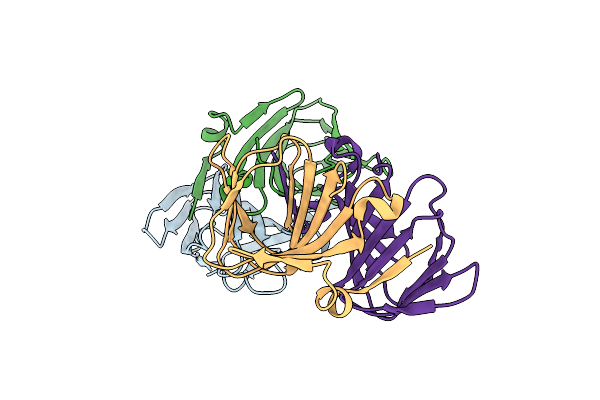 |
Crystal Structure Of The Ectoine Synthase From The Cold-Adapted Marine Bacterium Sphingopyxis Alaskensis
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-04-27 Classification: LYASE |
 |
High Resolution Structure Of The Ectoine Synthase From The Cold-Adapted Marine Bacterium Sphingopyxis Alaskensis
Organism: Sphingopyxis alaskensis (strain dsm 13593 / lmg 18877 / rb2256)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-04-27 Classification: LYASE Ligands: PGO |
 |
Organism: Sphingopyxis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-09-24 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of Ectd From S. Alaskensis With 2-Oxoglutarate And 5-Hydroxyectoine
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2014-09-10 Classification: OXIDOREDUCTASE Ligands: FE, AKG, 6CS |
 |
Organism: Sphingopyxis alaskensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-09-03 Classification: OXIDOREDUCTASE |

