Search Count: 267
All
Selected
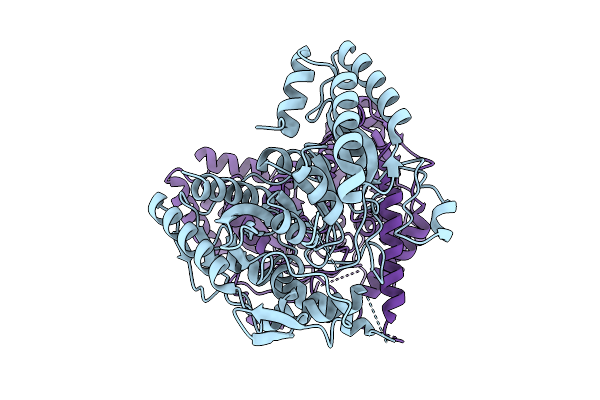 |
Organism: Sphingomonas sp. tpd3009
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2025-07-09 Classification: HYDROLASE |
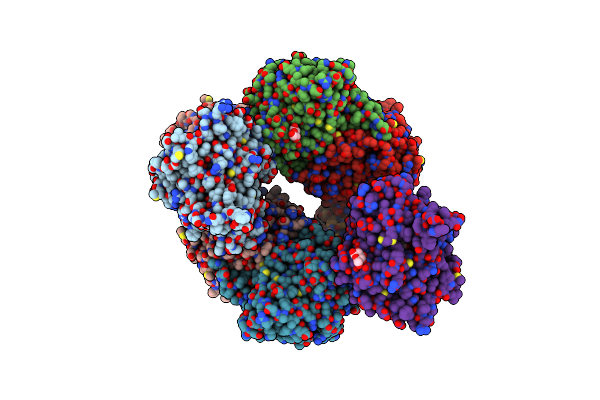 |
Organism: Sphingomonas sp. y57
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: ACT, BGC, ZN, K |
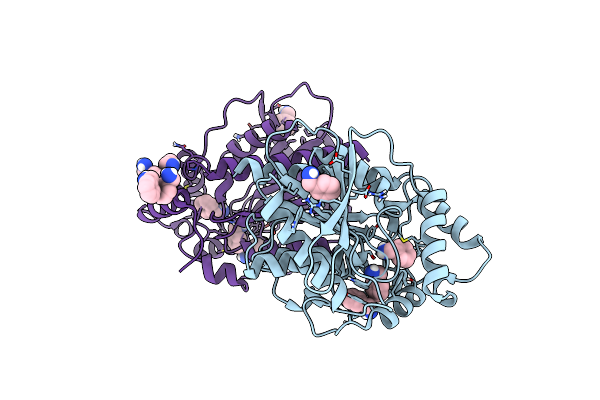 |
Crystal Structure Of The Lipase Spl From Sphingomonas Sp. Hxn-200 In Complex With Benzylamine
Organism: Sphingomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: ABN |
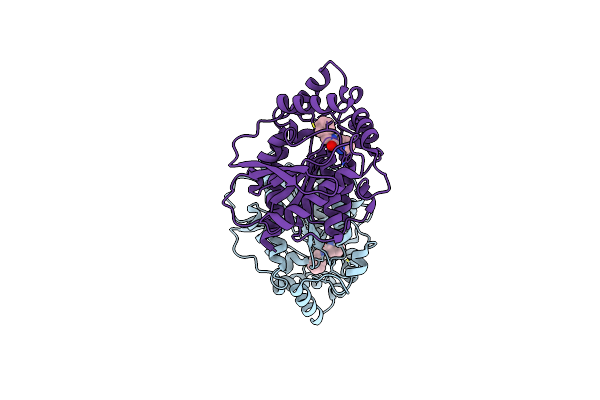 |
Crystal Structure Of The Lipase Spl From Sphingomonas Sp. Hxn-200 In Complex With N-Benzyl-Picolinamide
Organism: Sphingomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: XBW |
 |
Organism: Sphingomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: CL |
 |
Beta-Xylosidase Jb13Gh39P28 Showing Salt/Ethanol/Trypsin Tolerance, Low-Ph/Low-Temperature Activity, And Transformation Of Notoginsenosides
Organism: Sphingomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-24 Classification: HYDROLASE |
 |
Organism: Sphingomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-04-03 Classification: HYDROLASE |
 |
Crystal Structure Of Microcystinase C From Sphingomonas Sp. Acm-3962 At 2.6 A Resolution
Organism: Sphingomonas sp. acm-3962
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of L-2,4-Diketo-3-Deoxyrhamnonate Hydrolase From Sphingomonas Sp. (Apo-Form)
Organism: Sphingomonas sp. ska58
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-02-08 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of L-2,4-Diketo-3-Deoxyrhamnonate Hydrolase From Sphingomonas Sp. (Pyruvate Bound-Form)
Organism: Sphingomonas sp. ska58
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-02-08 Classification: HYDROLASE Ligands: MG, PYR |
 |
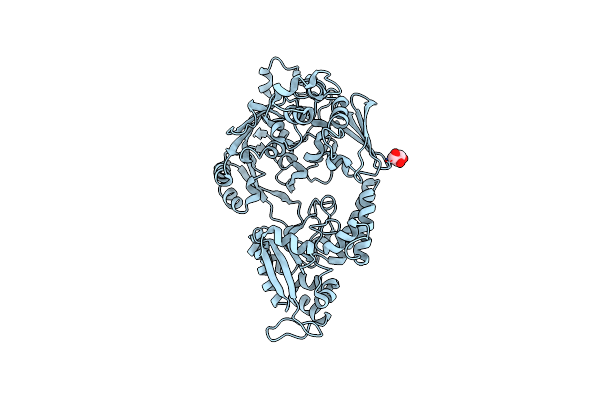
Crystal Structure Of Bacterial Chemotaxis-Dependent Pectin-Binding Protein Sph1118 In An Open Conformation
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-17 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
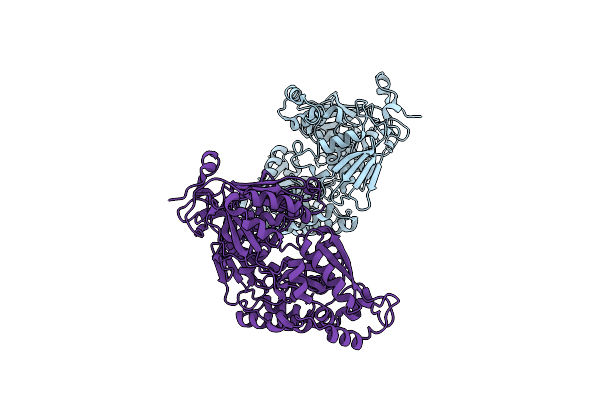
Crystal Structure Of Bacterial Chemotaxis-Dependent Pectin-Binding Protein Sph1118 In A Full Open Conformation
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-17 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
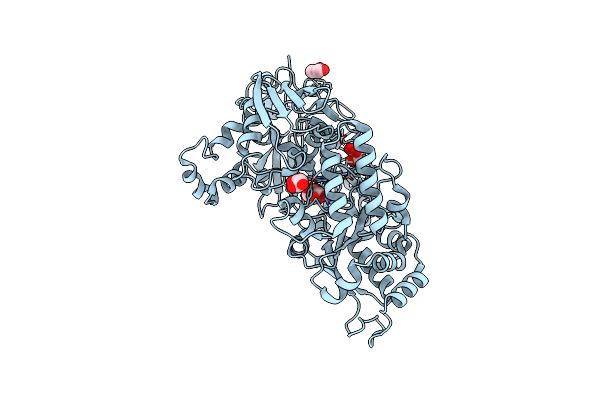
Crystal Structure Of Bacterial Chemotaxis-Dependent Pectin-Binding Protein Sph1118 In A Closed Conformation
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-08-17 Classification: SUGAR BINDING PROTEIN |
 |
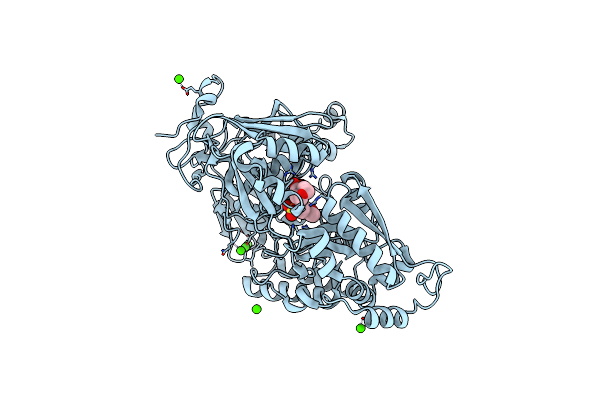
Crystal Structure Of Bacterial Chemotaxis-Dependent Pectin-Binding Protein Sph1118 In Complex With Galacturonic Acid
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-08-17 Classification: SUGAR BINDING PROTEIN Ligands: GOL, ADA |
 |
Crystal Structure Of Bacterial Chemotaxis-Dependent Pectin-Binding Protein Sph1118 In Complex With Mes
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-08-17 Classification: SUGAR BINDING PROTEIN Ligands: CA, MES |
 |
Crystal Structure Of Bacterial Chemotaxis-Dependent Pectin-Binding Protein Sph1118 In Complex With Unsaturated Trigalacturonic Acid
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-08-17 Classification: SUGAR BINDING PROTEIN Ligands: EPE, GOL, TLA |
 |
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-12-08 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-12-08 Classification: HYDROLASE Ligands: GD3 |
 |
A Structural Characterization Of Poly(Aspartic Acid) Hydrolase-1 From Sphingomonas Sp. Kt-1.
Organism: Sphingomonas sp. kt-1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-12-09 Classification: HYDROLASE |
 |
Crystal Structure Of Sphingomonas Sp. A1 Peroxidase Efeb Responsible For Import Of Iron
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-01-29 Classification: OXYGEN BINDING Ligands: HEM, OXY, PEG, EDO, PGE |

