Search Count: 34
 |
Organism: Sphaerobacter thermophilus (strain dsm 20745 / s 6022)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-12-21 Classification: HYDROLASE |
 |
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: QEO, MLI, PEG |
 |
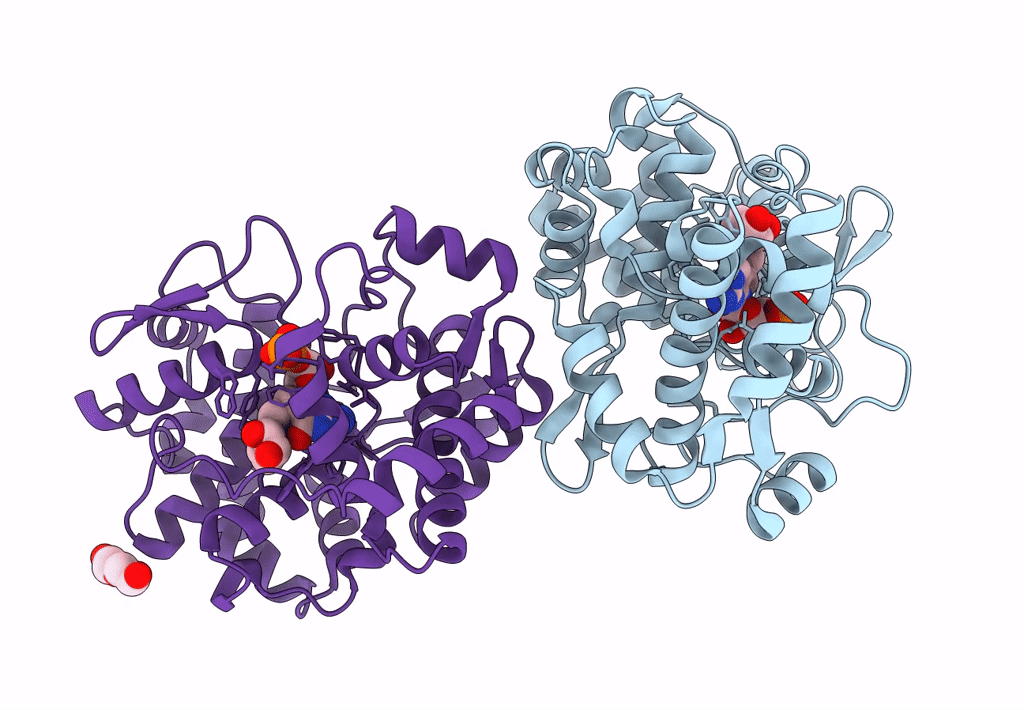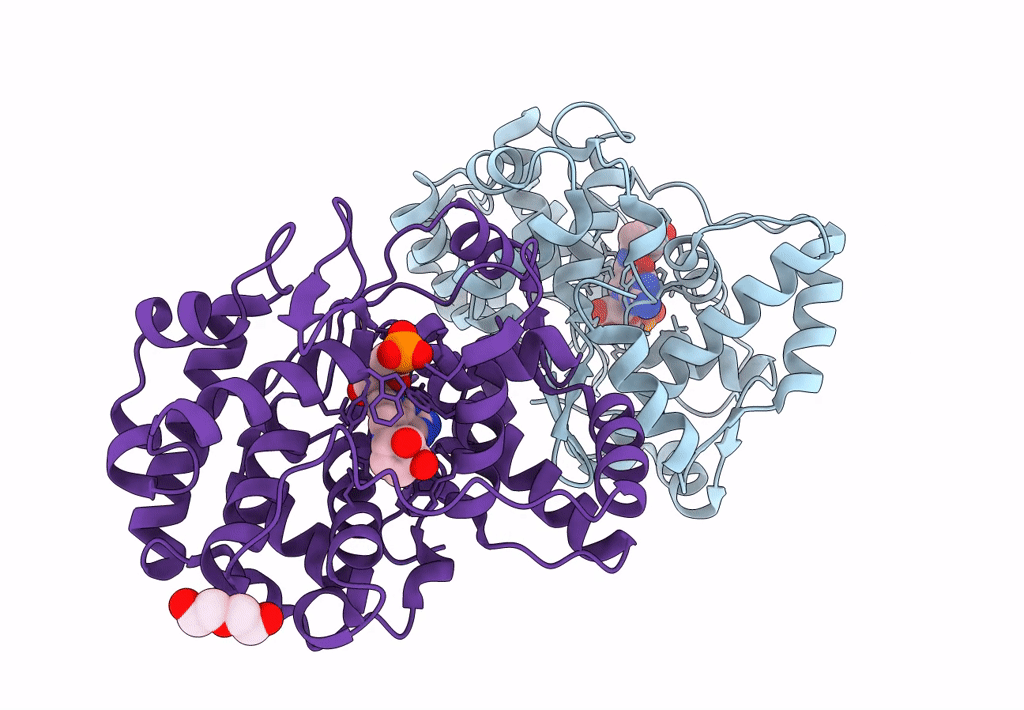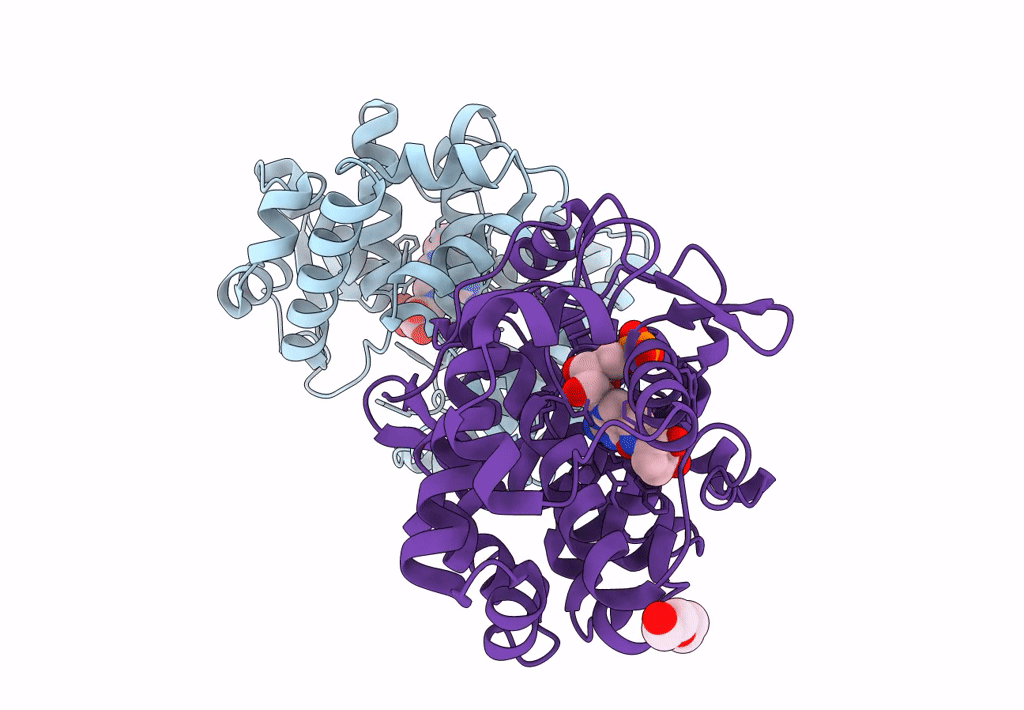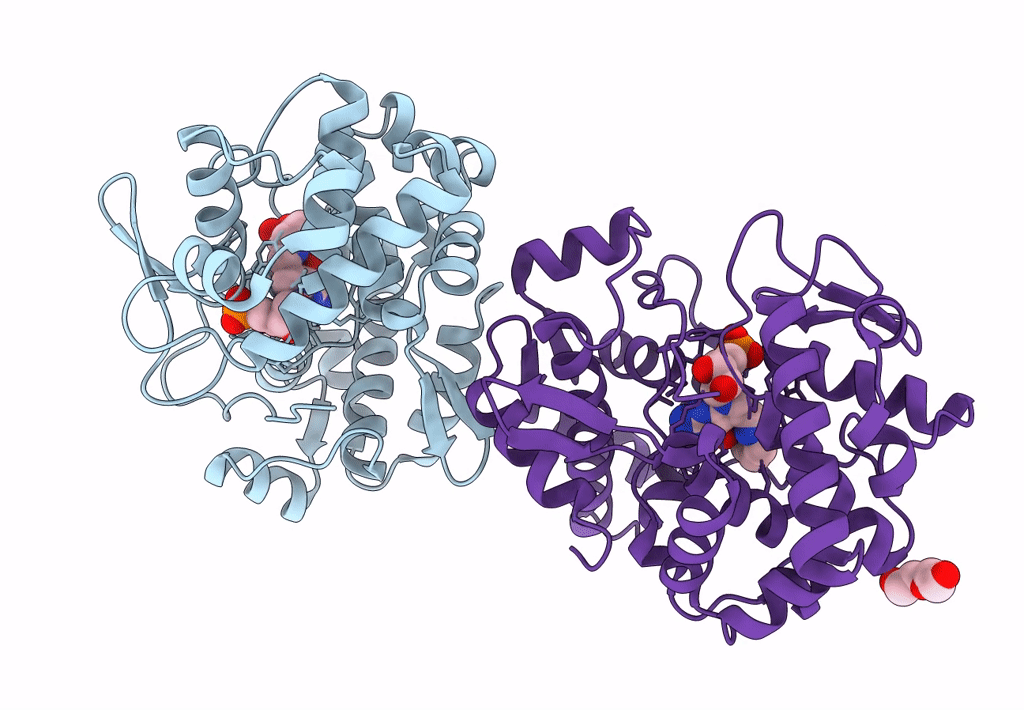
Crystal Structure Of Queuine Salvage Enzyme Duf2419 Mutant K199C, Complexed With Queuosine
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: QEO, MLI |
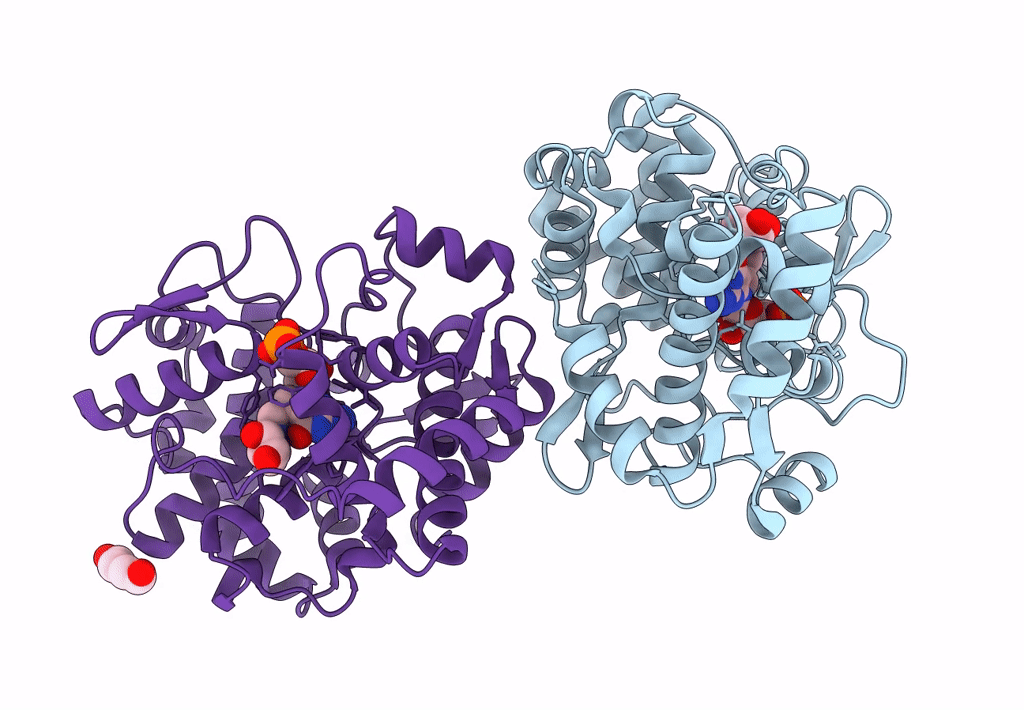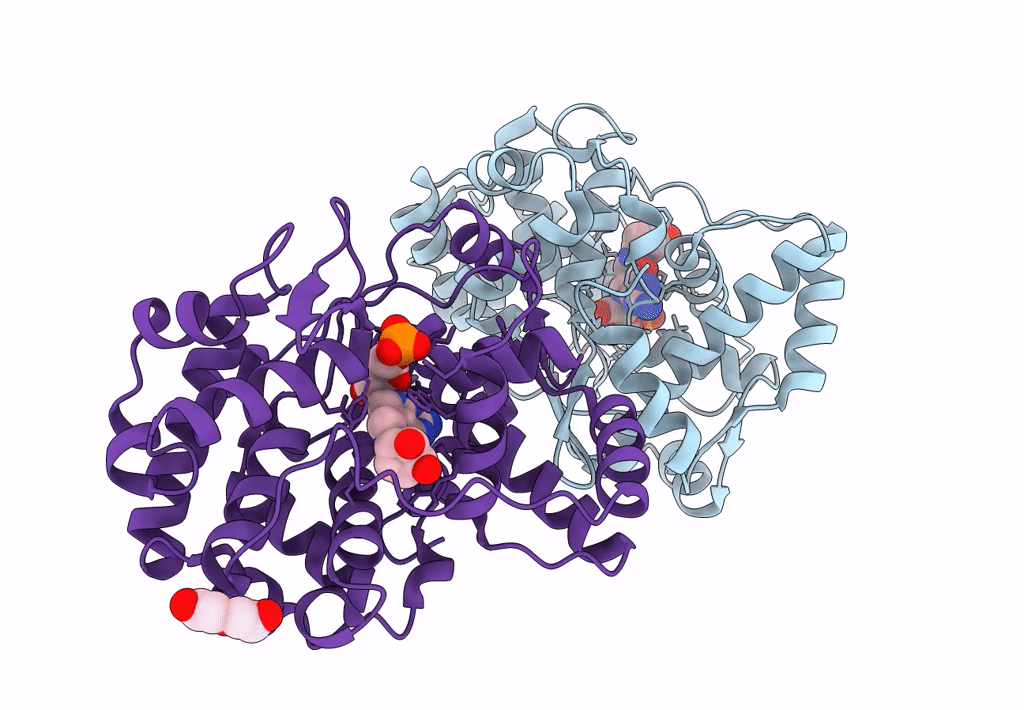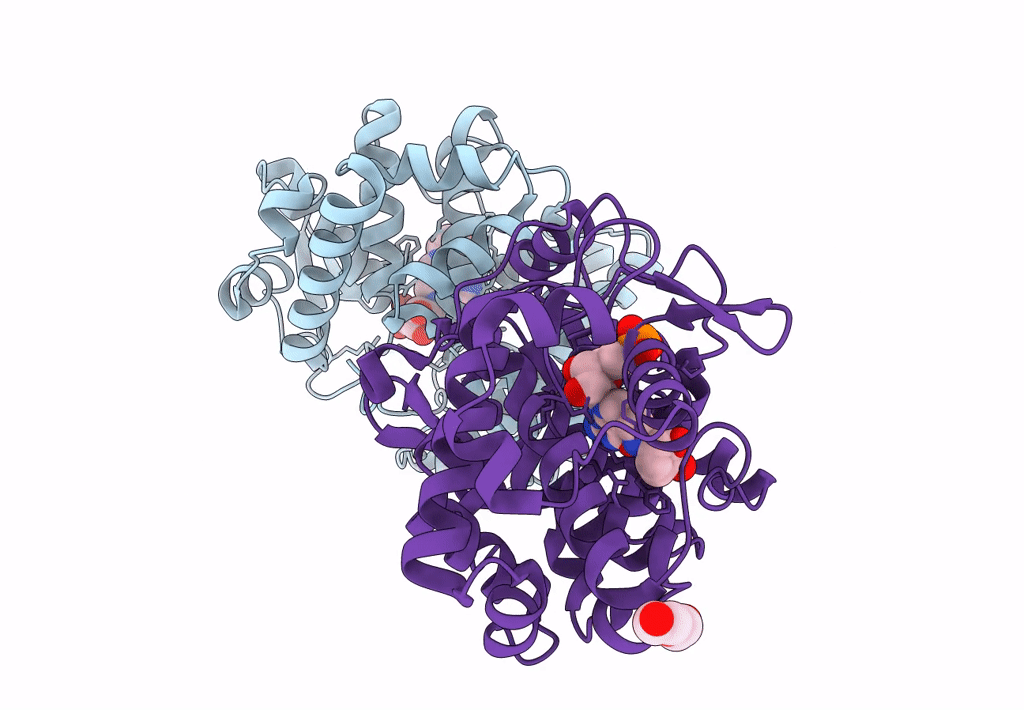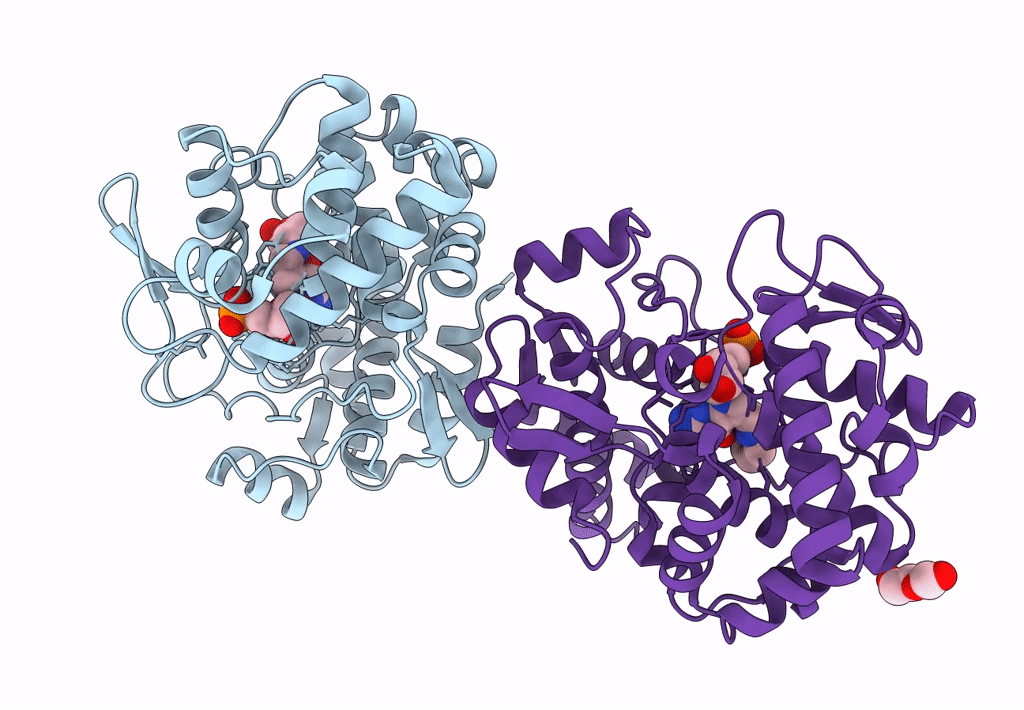 |
Crystal Structure Of Queuine Salvage Enzyme Duf2419, In Complex With Queuosine-5'-Monophosphate
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: 56B, NH4, PEG |
 |
Crystal Structure Of Queuine Salvage Enzyme Duf2419, Wild-Type (Non-His6X Tagged)
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-12-21 Classification: HYDROLASE |
 |
Crystal Structure Of Queuine Salvage Enzyme Duf2419 Mutant D231N, In Complex With Queuosine-5'-Monophosphate
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: 56B, PEG |
 |
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-09 Classification: TRANSFERASE Ligands: PLR |
 |
Crystal Structure Of A Radical Sam Methyltransferase From Sphaerobacter Thermophilus
Organism: Sphaerobacter thermophilus (strain dsm 20745 / s 6022)
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2019-04-10 Classification: TRANSFERASE Ligands: SF4, SAH, GOL, BR |
 |
Organism: Sphaerobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-10-26 Classification: UNKNOWN FUNCTION Ligands: GOL, CL |
 |
Crystal Structure Of Rimk Domain Protein Atp-Grasp From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-03-16 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
Organism: Sphaerobacter thermophilus (strain dsm 20745 / s 6022)
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2015-05-20 Classification: TRANSPORT PROTEIN Ligands: ZN, CL |
 |
The Crystal Structure Of Y333Q Mutant Pyridoxal-Dependent Decarboxylase From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-11-12 Classification: LYASE Ligands: PMP, NA, GOL |
 |
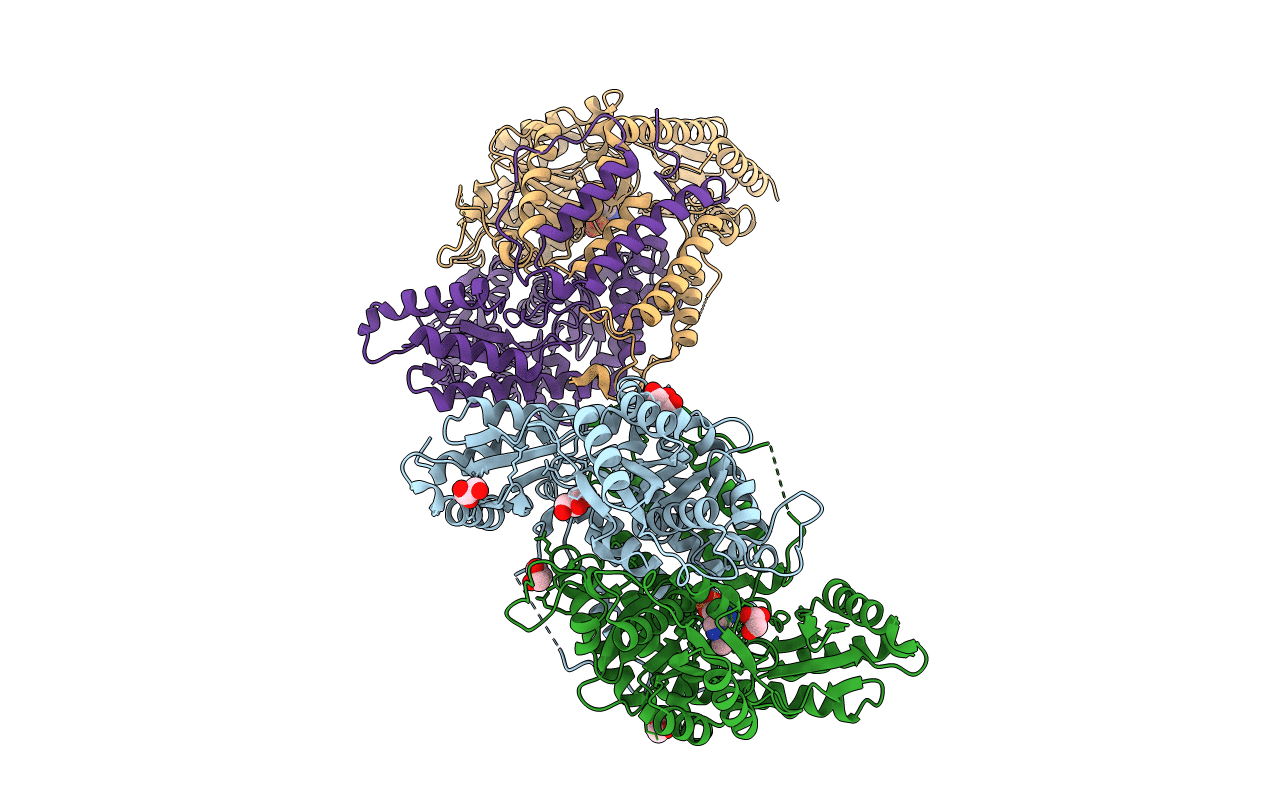
The Crystal Structure Of Y333N Mutant Pyridoxal-Dependent Decarboxylase From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-11-12 Classification: LYASE Ligands: GOL, PO4, PMP |
 |
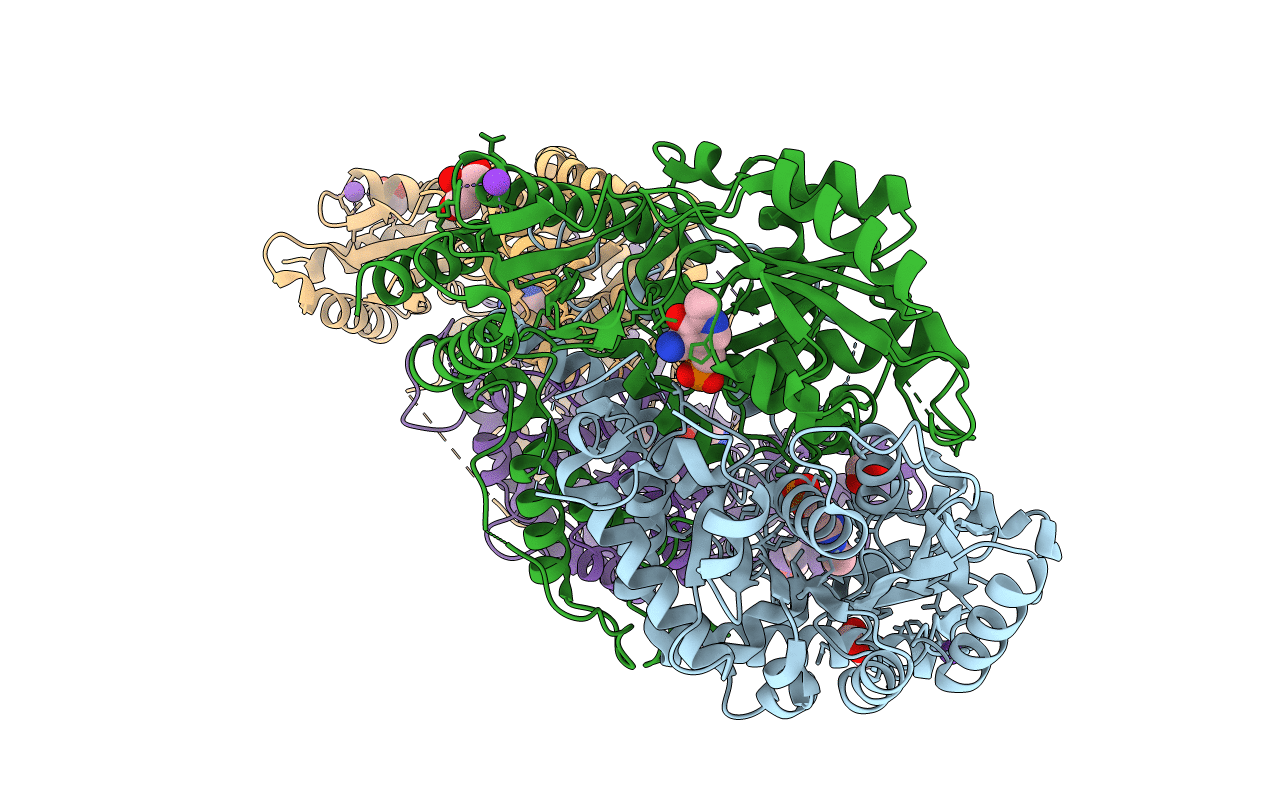
The Crystal Structure Of Y333Q Mutant Pyridoxal-Dependent Decarboxylase From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-11-12 Classification: LYASE Ligands: PMP, GOL, CL |
 |
The Clear Crystal Structure Of Pyridoxal-Dependent Decarboxylase From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-10-29 Classification: LYASE Ligands: PMP, GOL, K, ABU |
 |
The Yellow Crystal Structure Of Pyridoxal-Dependent Decarboxylase From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus dsm 20745
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-10-22 Classification: LYASE Ligands: CL, GOL, TRS |
 |
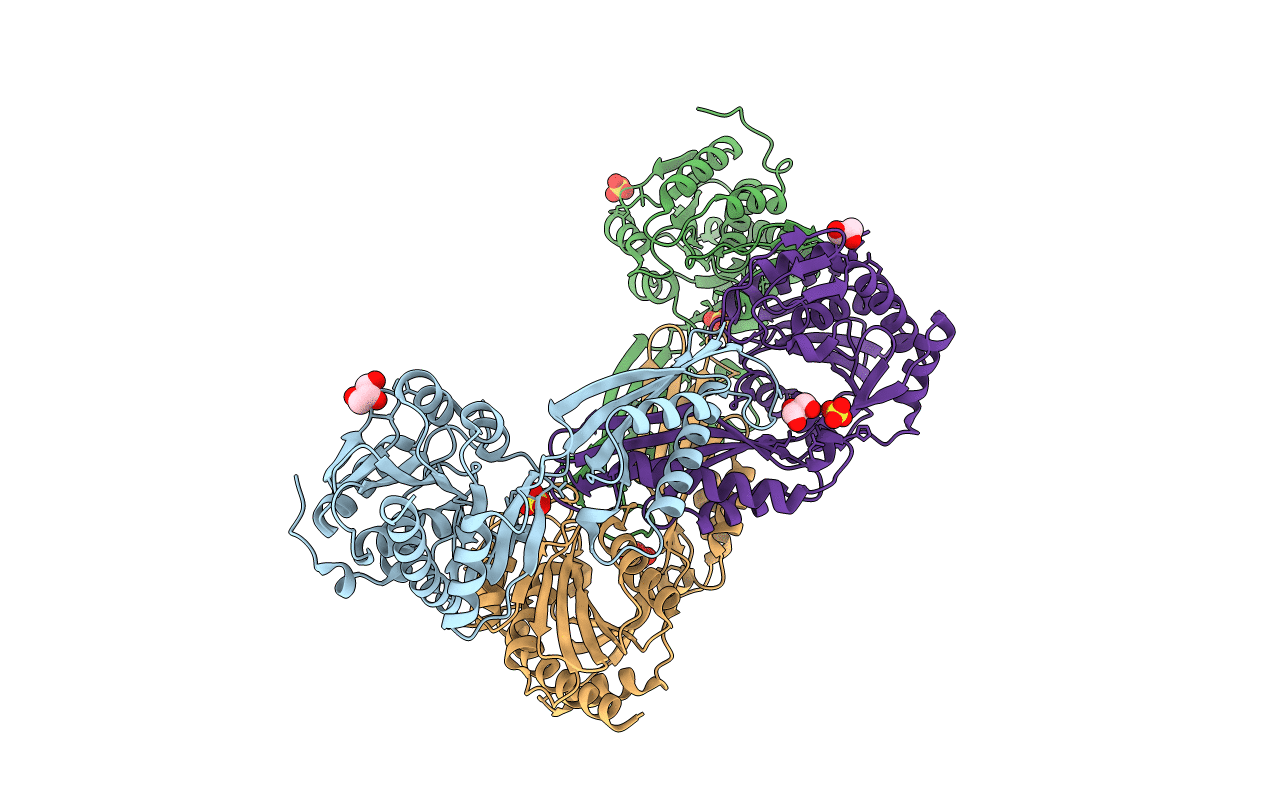
Crystal Structure Of Putative Thiolase From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-08-06 Classification: TRANSFERASE |
 |
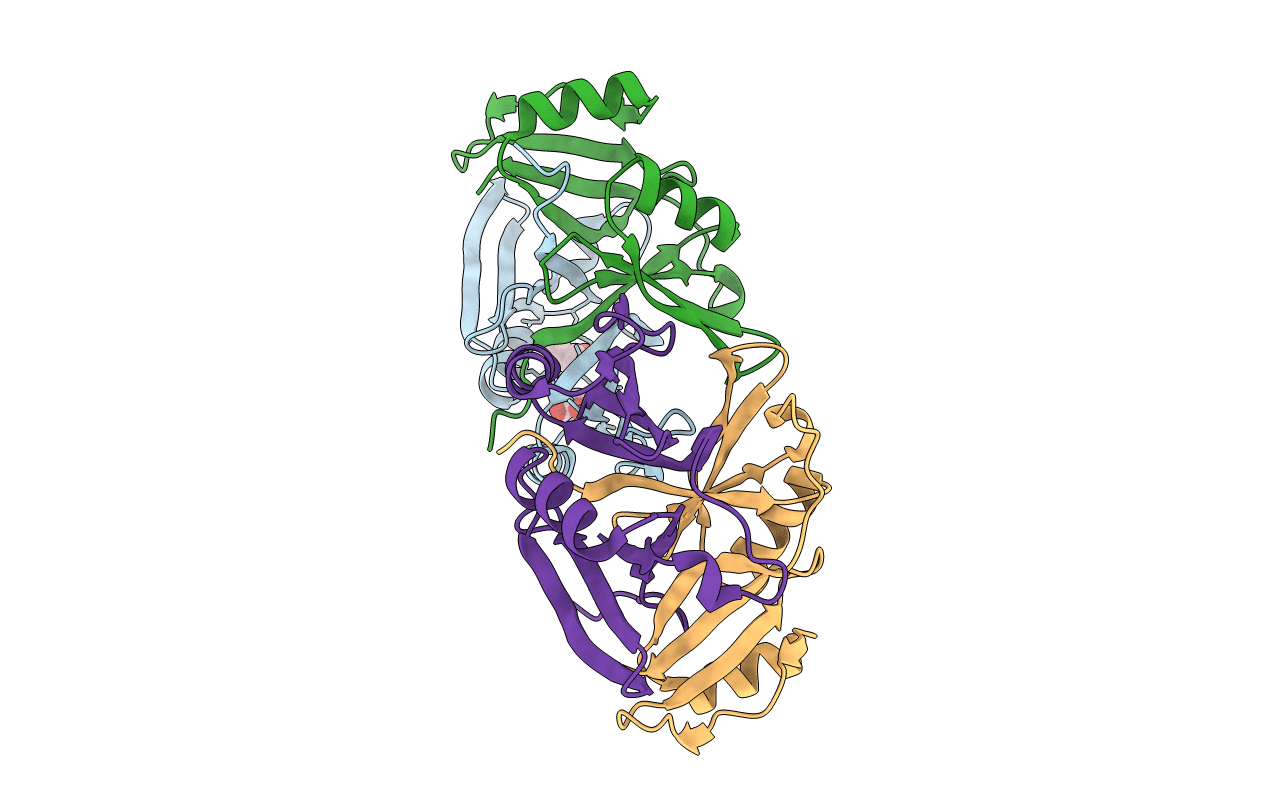
Crystal Structure Of N-Acetyl-Ornithine/N-Acetyl-Lysine Deacetylase From Sphaerobacter Thermophilus
Organism: Sphaerobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-07-02 Classification: HYDROLASE Ligands: SO4, GOL, CL |
 |
Crystal Structure Of Member Of Glyoxalase/Bleomycin Resistance Protein/Dioxygenase Superfamily From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-11-28 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
The Crystal Structure Of Beta-Lactamase From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-09-19 Classification: HYDROLASE Ligands: SO4, ACY |

