Search Count: 12
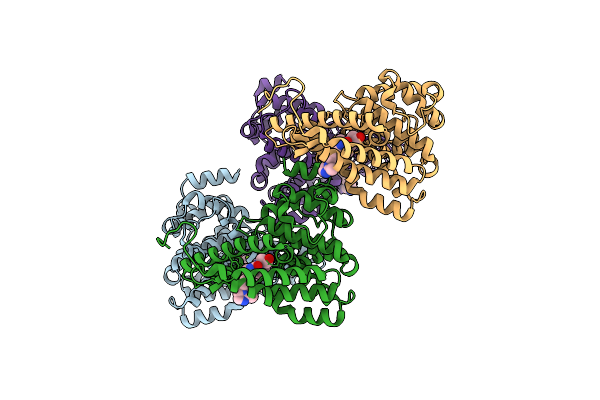 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2013-05-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, MG, 1L6 |
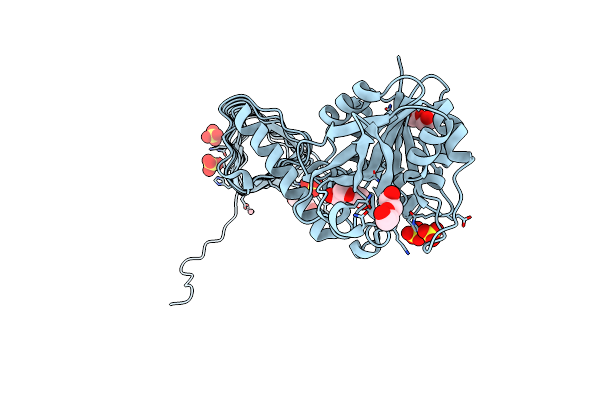 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: PEG, SO4, CO |
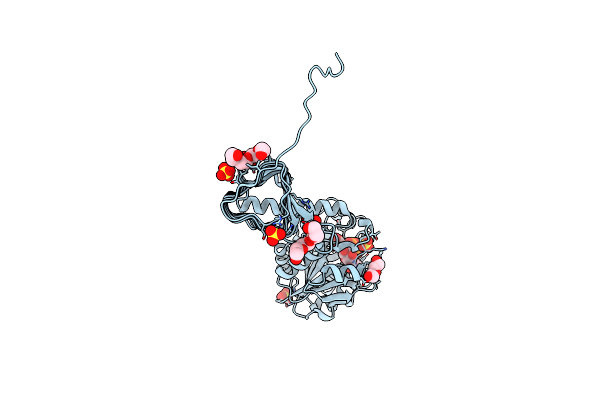 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: UD1, PG4, PEG, SO4 |
 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: H2U, MG, PG4, PGE, SO4 |
 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: UDP, PG4, PGE, SO4 |
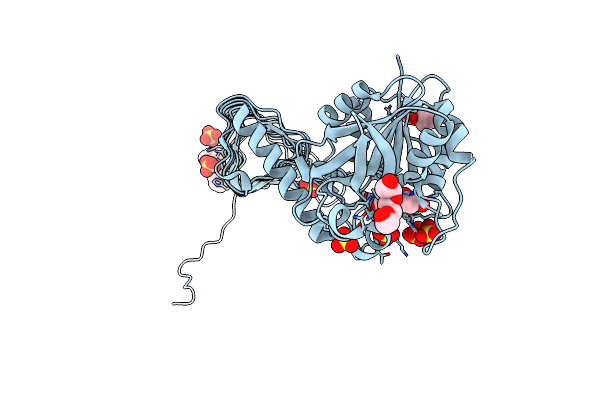 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: URI, PG4, PGE, SO4 |
 |
Phers From Staphylococcus Haemolyticus- Rational Protein Engineering And Inhibitor Studies
Organism: Staphylococcus haemolyticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-11-06 Classification: LIGASE Ligands: ZN, GAX |
 |
Phers From Staphylococcus Haemolyticus- Rational Protein Engineering And Inhibitor Studies
Organism: Staphylococcus haemolyticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-11-06 Classification: LIGASE Ligands: GAX, SO4, ZN |
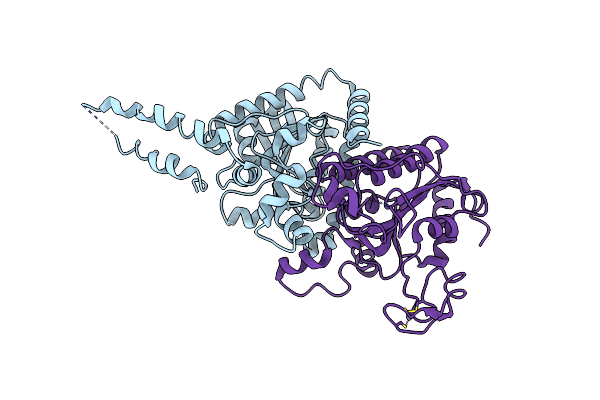 |
The Crystal Structure Of The Carboxyltransferase Subunit Of Acc From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2006-12-12 Classification: LIGASE Ligands: ZN |
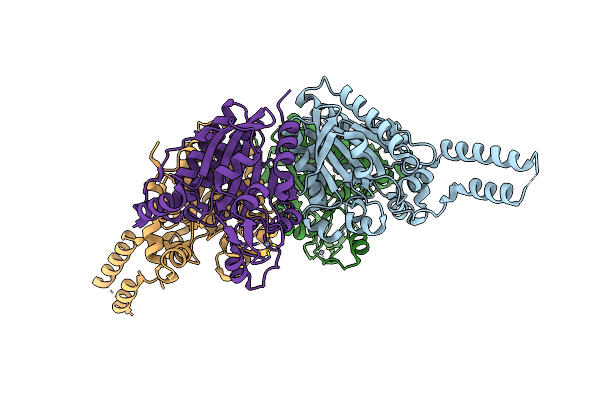 |
Crystal Structure Of The Carboxyltransferase Subunit Of Acc From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2006-12-05 Classification: TRANSFERASE Ligands: ZN |
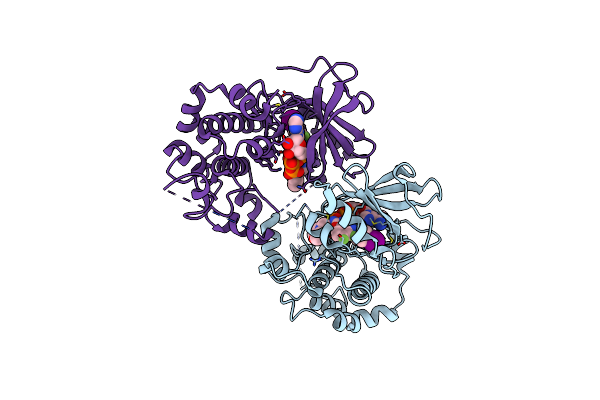 |
X-Ray Structure Of The Human Mitogen-Activated Protein Kinase Kinase 2 (Mek2)In A Complex With Ligand And Mgatp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2004-11-23 Classification: TRANSFERASE Ligands: MG, ATP, 5EA |
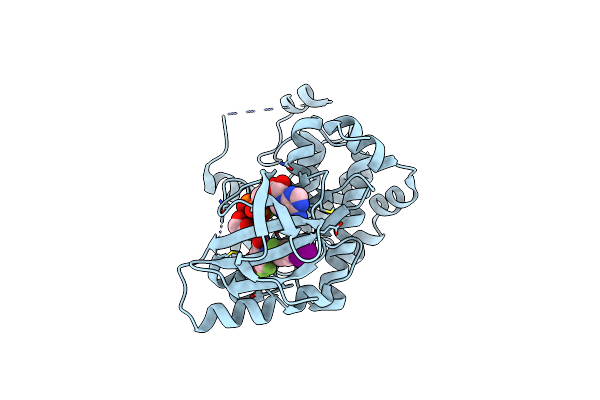 |
X-Ray Structure Of The Human Mitogen-Activated Protein Kinase Kinase 1 (Mek1) In A Complex With Ligand And Mgatp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2004-11-23 Classification: TRANSFERASE Ligands: MG, ATP, BBM |

