Search Count: 12
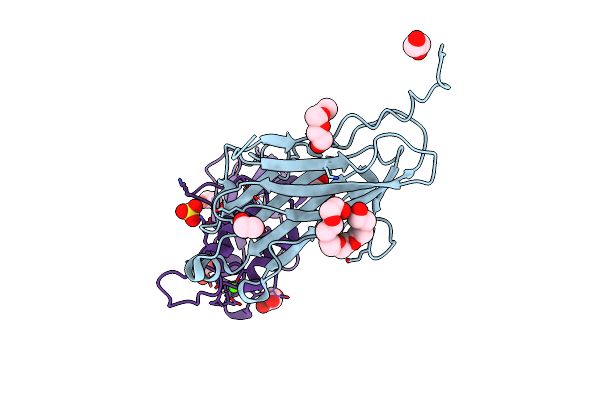 |
Crystal Structure Of The Cohscaa-Xdoccipb Type Ii Complex From Clostridium Thermocellum At 1.5Angstrom Resolution
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-09-06 Classification: CELL ADHESION Ligands: CA, SO4, GOL, EDO, PGE, P6G |
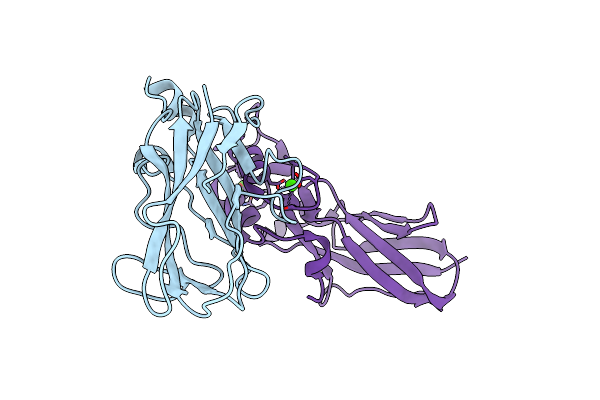 |
Crystal Structure Of The Cohscac2-Xdoccipa Type Ii Complex From Clostridium Thermocellum
Organism: Ruminiclostridium thermocellum ad2, Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-04-05 Classification: CARBOHYDRATE BINDING PROTEIN Ligands: CA |
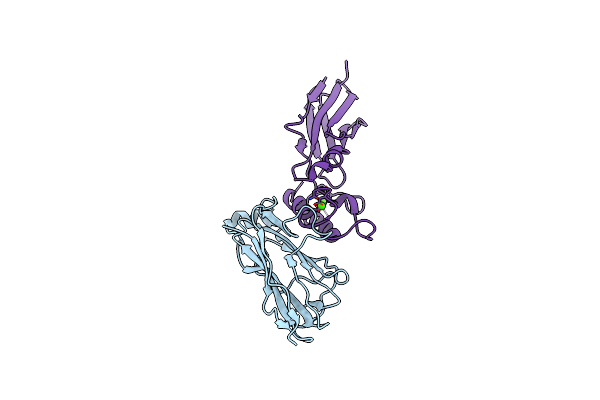 |
Organism: Ruminiclostridium thermocellum 27405, Clostridium thermocellum 27405
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-03-29 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Organism: Schizophyllum commune h4-8
Method: SOLUTION NMR Release Date: 2016-03-16 Classification: PROTEIN FIBRIL |
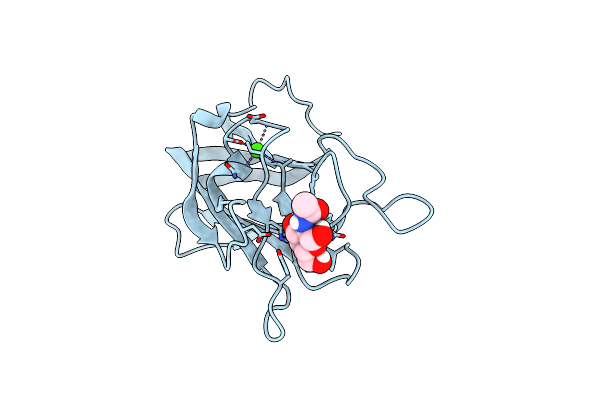 |
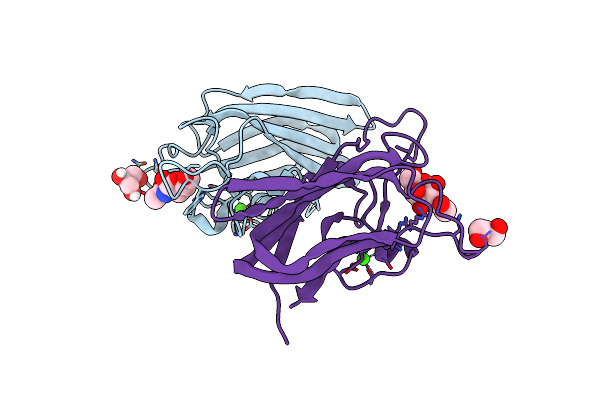
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens In Complex With N-Acetylgalactosamine
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-10-07 Classification: SUGAR BINDING PROTEIN Ligands: NGA, CA |
 |
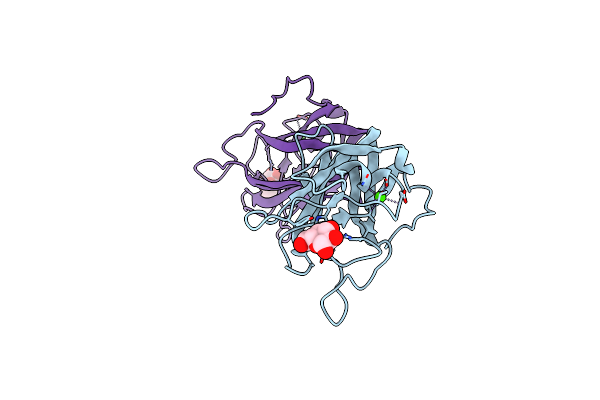
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-10-07 Classification: SUGAR BINDING PROTEIN Ligands: NGA, CA, GOL |
 |
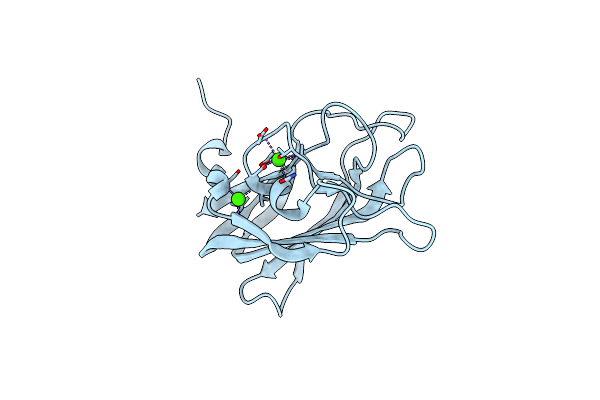
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens In Complex With Galactose
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-24 Classification: SUGAR BINDING PROTEIN Ligands: CA, GAL |
 |
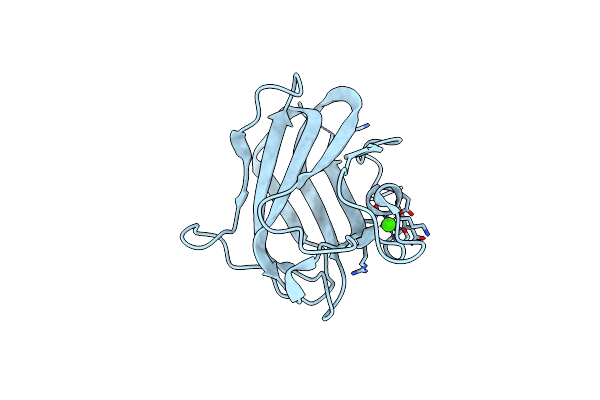
Structure Of Cbm32-3 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2014-07-23 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Structure Of Cbm32-1 From A Family 31 Glycoside Hydrolase From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-07-16 Classification: SUGAR BINDING PROTEIN Ligands: CA, MG |
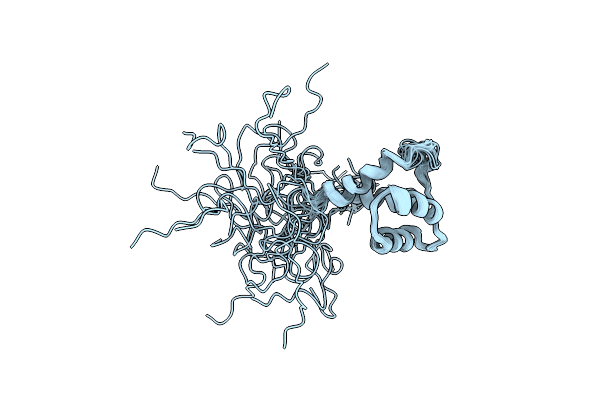 |
Solution Structure Of The Small Dictyostelium Discoideium Myosin Light Chain Mlcb Provides Insights Into Iq-Motif Recognition Of Class I Myosin Myo1B
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2013-12-11 Classification: CONTRACTILE PROTEIN |
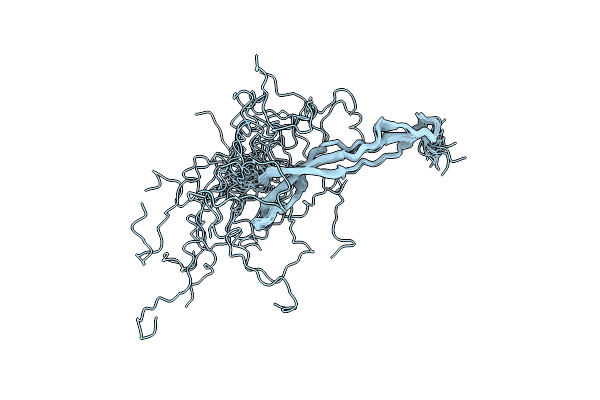 |
Conformational Analysis Of Strh, The Surface-Attached Exo- Beta-D-N-Acetylglucosaminidase From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae
Method: SOLUTION NMR Release Date: 2012-11-28 Classification: HYDROLASE |
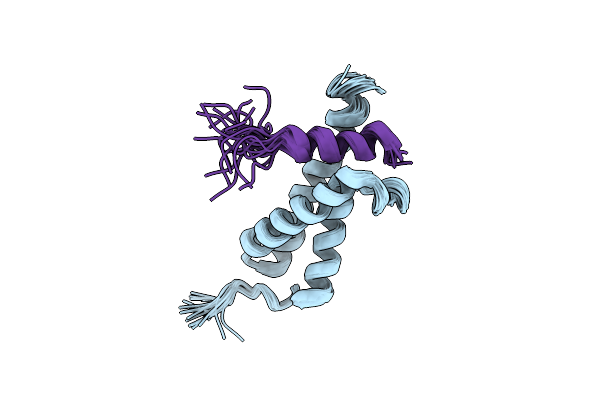 |
The Structure Of E-Protein Activation Domain 1 Bound To The Kix Domain Of Cbp/P300 Elucidates Leukemia Induction By E2A-Pbx1
|

