Search Count: 14
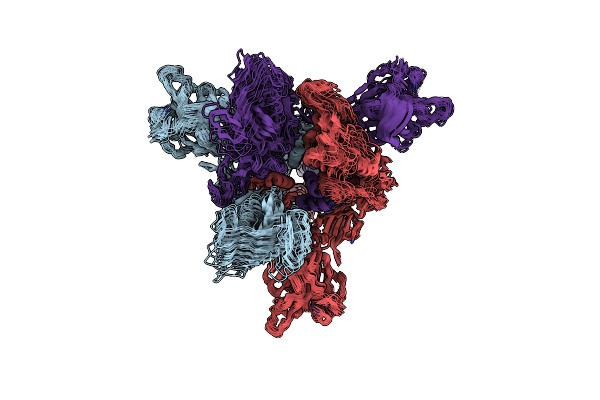 |
Sars-Cov-2 Spike Protein Beta Variant At 4C Structural Flexibility / Heterogeneity Analyses
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: VIRAL PROTEIN |
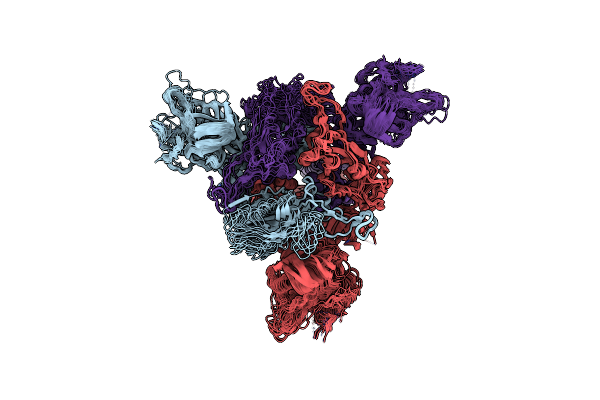 |
Sars-Cov-2 Spike Protein Beta Variant At 37C Structural Flexibility / Heterogeneity Analyses
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Shiga Toxin 2 In Complex With The Native Ribosomal P-Stalk
Organism: Shigella dysenteriae, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: TOXIN |
 |
The Structure Of Photosystem I Tetramer From Chroococcidiopsis Ts-821, A Thermophilic, Unicellular, Non-Heterocyst-Forming Cyanobacterium
Organism: Chroococcidiopsis sp. ts-821
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: PHOTOSYNTHESIS Ligands: LHG, CLA, PQN, BCR, SF4 |
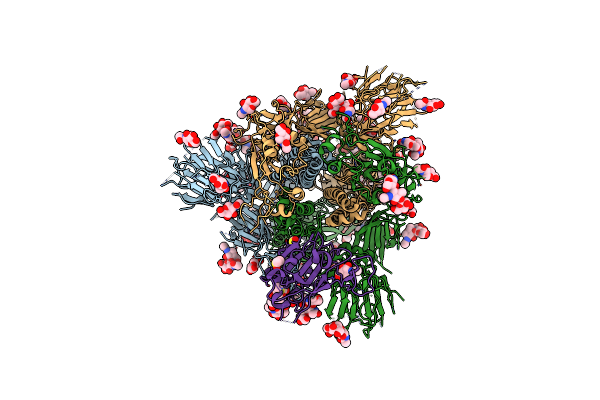 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: VIRAL PROTEIN Ligands: NAG, MAN, DMS |
 |
Sars-Cov-2 Spike In Prefusion State (Flexibility Analysis, 1-Up Closed Conformation)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: VIRAL PROTEIN Ligands: NAG, MAN, DMS |
 |
Sars-Cov-2 Spike In Prefusion State (Flexibility Analysis, 1-Up Open Conformation)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: VIRAL PROTEIN Ligands: NAG, MAN, DMS |
 |
Influenza A Nucleoprotein Docked Into 3D Helical Structure Of The Wild Type Ribonucleoprotein Complex Obtained Using Cryoem. Conformation 3.
Organism: Influenza a virus (a/wilson-smith/1933(h1n1))
Method: ELECTRON MICROSCOPY Release Date: 2020-02-19 Classification: VIRAL PROTEIN |
 |
Influenza A Nucleoprotein Docked Into 3D Helical Structure Of The Wild Type Ribonucleoprotein Complex Obtained Using Cryoem. Conformation 1.
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1), Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2020-02-12 Classification: VIRAL PROTEIN |
 |
Influenza A Nucleoprotein Docked Into 3D Helical Structure Of The Wild Type Ribonucleoprotein Complex Obtained Using Cryoem. Conformation 4.
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2020-02-12 Classification: VIRAL PROTEIN |
 |
Influenza A Nucleoprotein Docked Into The 3D Helical Structure Of The Wild Type Ribonucleoprotein Complex Obtained Using Cryoem. Conformation 5.
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: ELECTRON MICROSCOPY Release Date: 2020-01-29 Classification: VIRAL PROTEIN |
 |
Influenza A Nucleoprotein Docked Into 3D Helical Structure Of The Wild Type Ribonucleoprotein Complex Obtained Using Cryoem. Conformation 2.
Organism: Influenza a virus (a/wilson-smith/1933(h1n1))
Method: ELECTRON MICROSCOPY Release Date: 2019-11-13 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2018-08-08 Classification: CHAPERONE Ligands: MG, EDO, PEG |
 |
Variable Internal Flexibility Characterizes The Helical Capsid Formed By Agrobacterium Vire2 Protein On Single-Stranded Dna.
Organism: Agrobacterium tumefaciens
Method: ELECTRON MICROSCOPY Resolution:20.00 Å Release Date: 2013-06-26 Classification: DNA BINDING PROTEIN |

