Search Count: 16
 |
The Structure Of The Catalytic Domain Of Afaa11B From The Filamentous Fungus Aspergillus Fumigatus
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, ASC, NAG, SO4 |
 |
Crystal Structure Of Aspergillus Fumigatus Aa11 Lytic Polysaccharide Monooxygenase (Afaa11B) In Complex With Cu(Ii)
Organism: Aspergillus fumigatus af293
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-04-23 Classification: METAL BINDING PROTEIN Ligands: SO4, NAG, CU |
 |
A Gh18 From Haloalkaliphilic Bacterium Unveils Environment-Dependent Variations In The Catalytic Machinery Of Chitinases
Organism: Chitinivibrio alkaliphilus acht1
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-01-17 Classification: HYDROLASE Ligands: EDO, NA |
 |
A Gh18 From Haloalkaliphilic Bacterium Unveils Environment-Dependent Variations In The Catalytic Machinery Of Chitinases
Organism: Chitinivibrio alkaliphilus acht1
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2024-01-17 Classification: HYDROLASE Ligands: EDO |
 |
High Resolution Structure Of The Iron Superoxide Dismutase From Thermobifida Fusca
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2023-09-27 Classification: ELECTRON TRANSPORT Ligands: FE, BO3 |
 |
Structural And Functional Characterization Of The Bacterial Lytic Polysaccharide Monooxygenase Sclpmo10D
Organism: Streptomyces coelicolor a3(2)
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2023-04-19 Classification: OXIDOREDUCTASE Ligands: CU, SO4, NA |
 |
Cu(I) Nmr Solution Structure Of The Chitin-Active Lytic Polysaccharide Monooxygenase Bllpmo10A
Organism: Bacillus licheniformis dsm 13 = atcc 14580
Method: SOLUTION NMR Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: CU1 |
 |
Nmr Solution Structure Of The Family 14 Carbohydrate Binding Module (Cbm14) From Human Chitotriosidase
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2020-01-15 Classification: SUGAR BINDING PROTEIN |
 |
Structural And Thermodynamic Signatures Of Ligand Binding To An Enigmatic Chitinase-D From Serratia Proteamaculans
Organism: Serratia proteamaculans (strain 568)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2019-03-06 Classification: HYDROLASE Ligands: AO3, EDO |
 |
Key Residues Affecting Transglycosylation Activity In Family 18 Chitinases - Insights Into Donor And Acceptor Subsites
Organism: Serratia proteamaculans (strain 568)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-07-04 Classification: HYDROLASE Ligands: EDO, CL |
 |
Nmr Solution Structure Of The Apo-Form Of The Chitin-Active Lytic Polysaccharide Monooxygenase Bllpmo10A
Organism: Bacillus licheniformis
Method: SOLUTION NMR Release Date: 2017-10-25 Classification: OXIDOREDUCTASE |
 |
The Structure Of The Catalytic Domain Of Nclpmo9C From The Filamentous Fungus Neurospora Crassa
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2015-07-22 Classification: OXIDOREDUCTASE Ligands: CU, GOL |
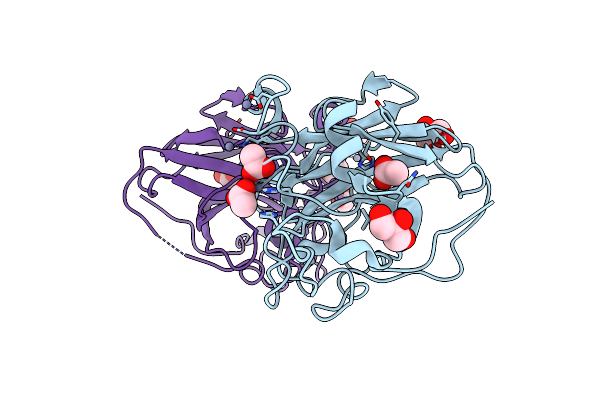 |
The Structure Of The Catalytic Domain Of Nclpmo9C From The Filamentous Fungus Neurospora Crassa
Organism: Neurospora crassa (strain atcc 24698 / 74-or23-1a / cbs 708.71 / dsm 1257 / fgsc 987)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-07-22 Classification: OXIDOREDUCTASE Ligands: ZN, ACT, GOL |
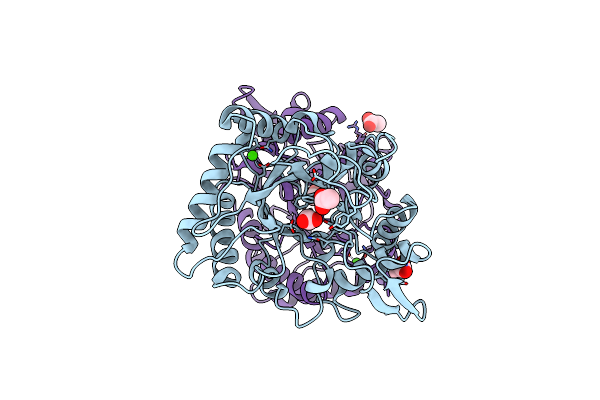 |
Hallmarks Of Processive And Non-Processive Glycoside Hydrolases Revealed From Computational And Crystallographic Studies Of The Serratia Marcescens Chitinases
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2012-09-05 Classification: HYDROLASE Ligands: CA, ACT |
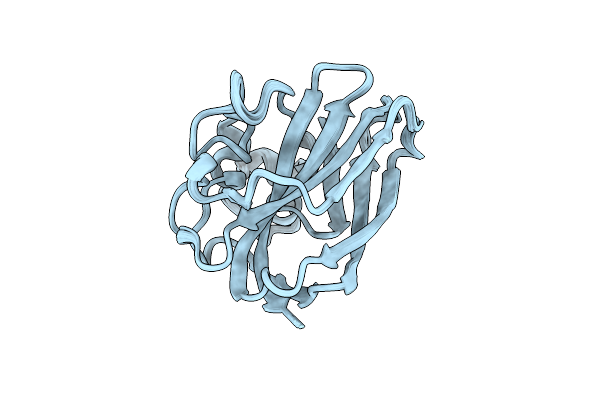 |
Organism: Serratia marcescens
Method: SOLUTION NMR Release Date: 2011-08-31 Classification: Chitin Binding Protein |
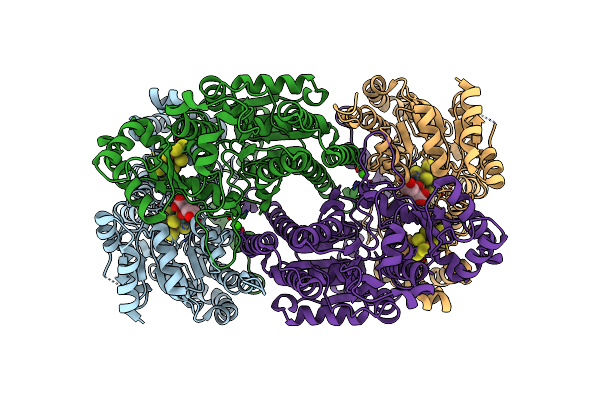 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2002-11-20 Classification: OXIDOREDUCTASE Ligands: HCA, CFM, CLP, CA |

