Search Count: 14
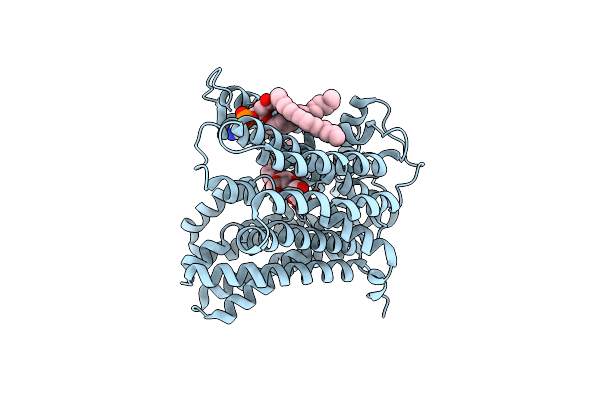 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: LMT, PEE |
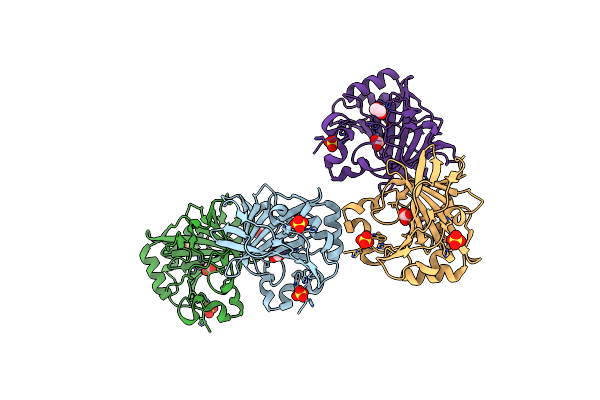 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-04 Classification: LYASE Ligands: SO4, ZN, BCT, EDO |
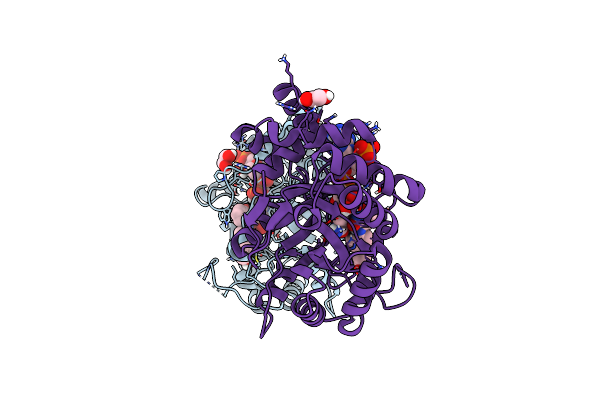 |
Structure Of Aldo-Keto Reductase 1C3 (Akr1C3) In Complex With An Inhibitor M689, With The 3-Hydroxy-Benzoisoxazole Moiety. Resolution 2.0A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-06 Classification: OXIDOREDUCTASE Ligands: NAP, YMC, EDO |
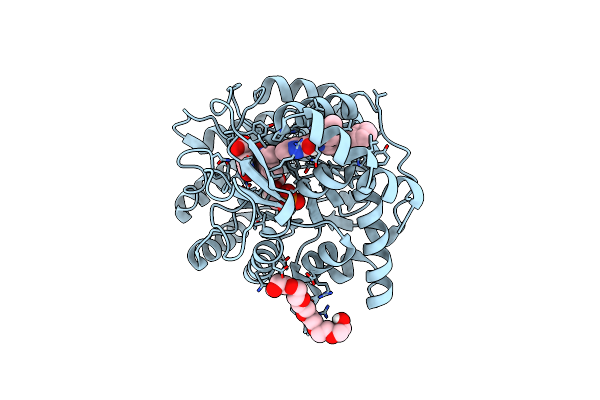 |
Crystal Structure Of Human Dihydroorotate Dehydrogenase In Complex With The Inhibitor 2-Hydroxy-N-(2-Ispropyl-5-Methyl-4-Phenoxyphenyl)Pyrazolo[1,5-A]Pyridine-3-Carboxamide.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-10-12 Classification: OXIDOREDUCTASE Ligands: FMN, ORO, IEQ, P6G, ACT |
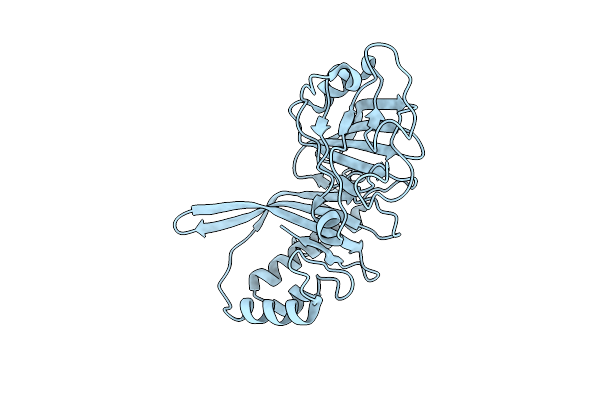 |
Crystal Structure Of The Helical Cell Shape Determining Protein Pgp2 From Campylobacter Jejuni
Organism: Campylobacter jejuni subsp. jejuni
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-03-17 Classification: HYDROLASE |
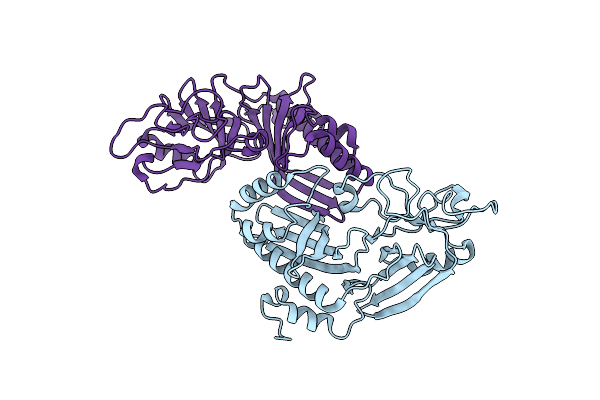 |
Crystal Structure Of The Helical Cell Shape Determining Protein Pgp2 (K307A Mutant) From Campylobacter Jejuni
Organism: Campylobacter jejuni subsp. jejuni
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-03-17 Classification: HYDROLASE |
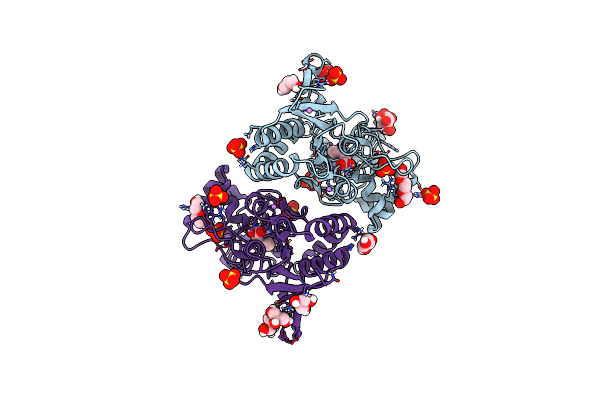 |
Structure Of Glua2 Ligand-Binding Domain (S1S2J) In Complex With The Agonist (S)-2-Amino-3-(1-Ethyl-4-Hydroxy-1H-1,2,3-Triazol-5-Yl)Propanoic Acid At 1.4 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-04-17 Classification: MEMBRANE PROTEIN Ligands: SO4, HJ8, GOL, PGE, CL, CIT, LI, PEG |
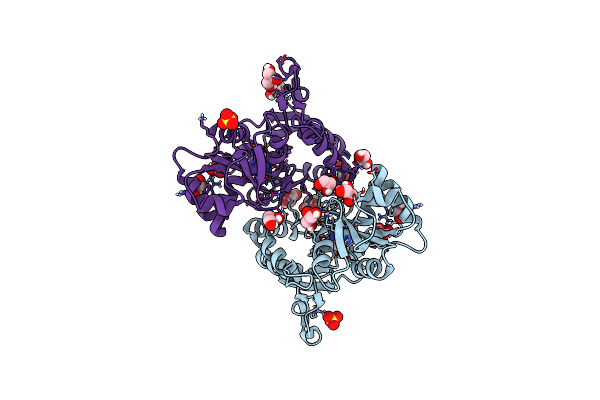 |
Structure Of Glua2 Ligand-Binding Domain (S1S2J) In Complex With The Agonist (S)-2-Amino-3-(2-Methyl-5-Hydroxy-2H-1,2,3-Triazol-4-Yl)Propanoic Acid At 1.55 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-04-17 Classification: MEMBRANE PROTEIN Ligands: HJH, SO4, GOL, CL, LI |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2010-01-19 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2009-08-11 Classification: HYDROLASE Ligands: FMT, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-02-17 Classification: TRANSCRIPTION REGULATOR/DNA Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-01-20 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2007-09-04 Classification: DNA BINDING PROTEIN Ligands: CO, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-06-12 Classification: ANTITUMOR PROTEIN Ligands: SO4, CL, EDO |

