Search Count: 44
 |
Crystal Structure Of Apo-Form Artificial Metalloprotein Incorporating A Tp Ligand
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-10-15 Classification: METAL BINDING PROTEIN |
 |
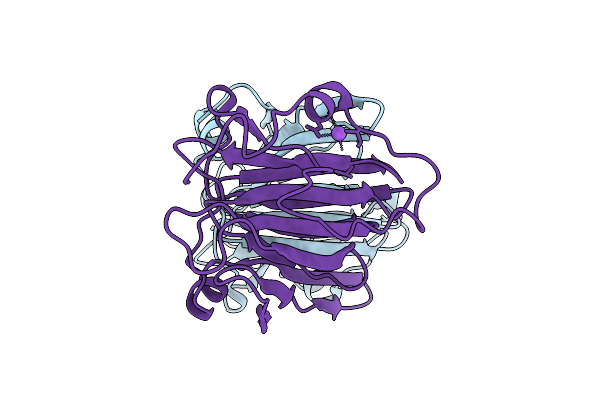
Crystal Structure Of Cu-Bound Artificial Metalloprotein Incorporating A Tp Ligand
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2025-10-15 Classification: METAL BINDING PROTEIN Ligands: CU, NA, SO4 |
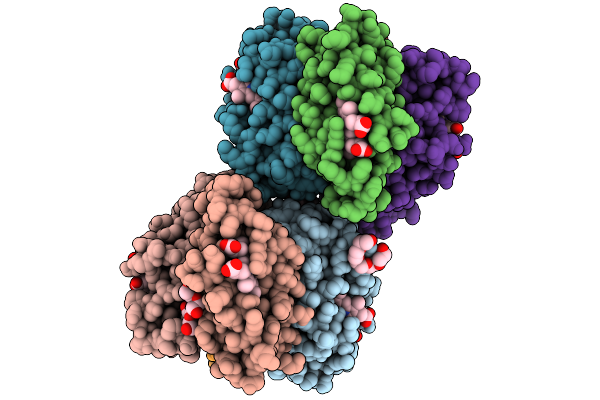 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-09-10 Classification: ELECTRON TRANSPORT Ligands: HEC, ZN, PEG, 1PE |
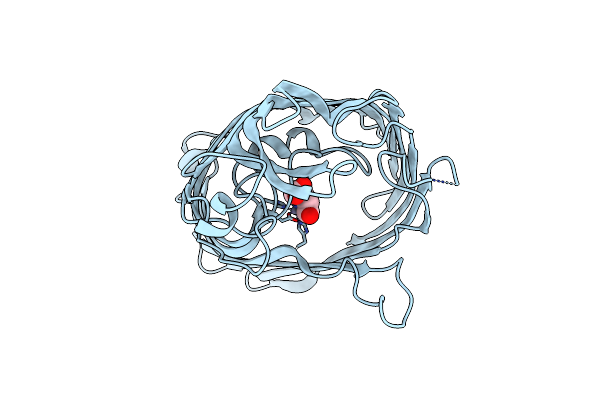 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: ZN, BCN |
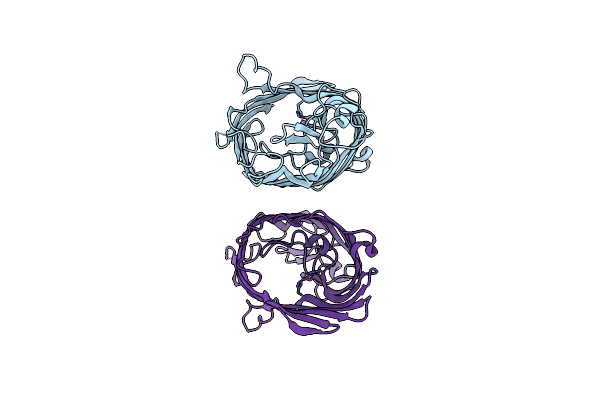 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: MN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2025-07-02 Classification: ISOMERASE Ligands: CU |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-07-02 Classification: ISOMERASE Ligands: ZN |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-07-02 Classification: ISOMERASE Ligands: ZN |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-07-02 Classification: ISOMERASE Ligands: MN |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-07-02 Classification: ISOMERASE Ligands: FE |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-07-02 Classification: ISOMERASE Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-07-02 Classification: ISOMERASE Ligands: CU |
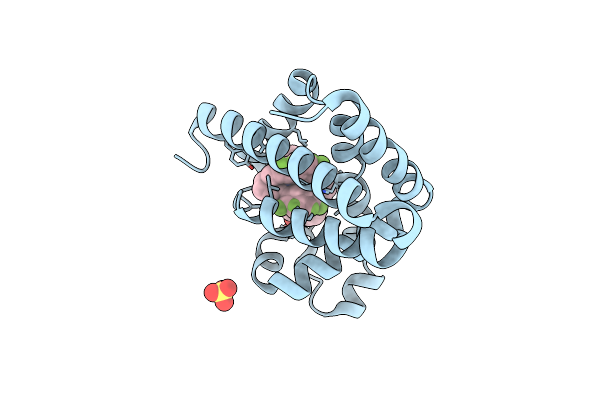 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-03-15 Classification: OXYGEN STORAGE Ligands: KKC, IRQ, SO4 |
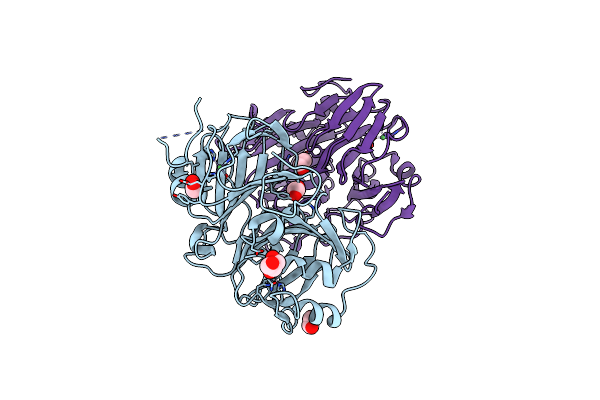 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-03-08 Classification: OXIDOREDUCTASE Ligands: EDO, GOL, NI |
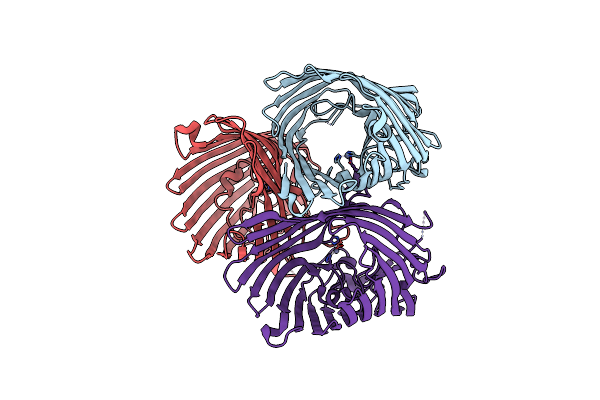 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: ZN |
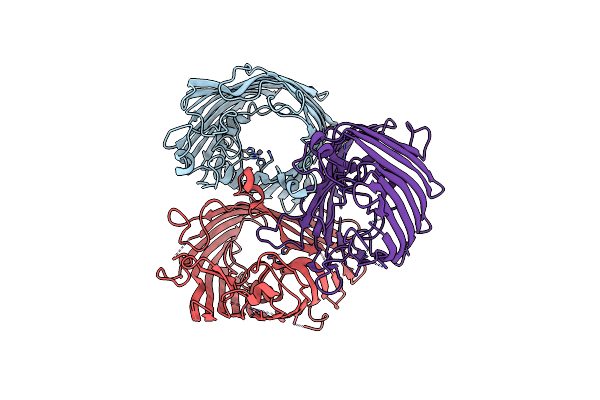 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: ZN |

