Search Count: 183
 |
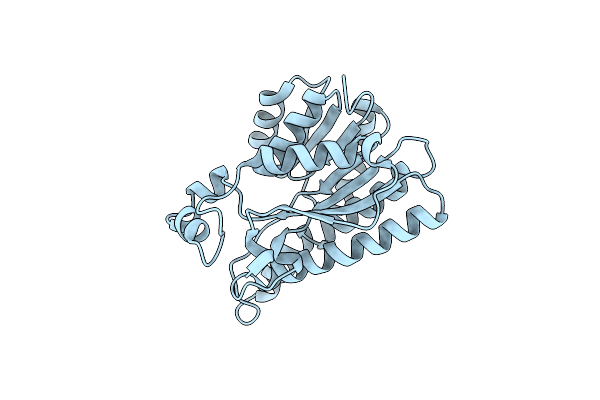
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: HYDROLASE Ligands: GOL, CA, TLA |
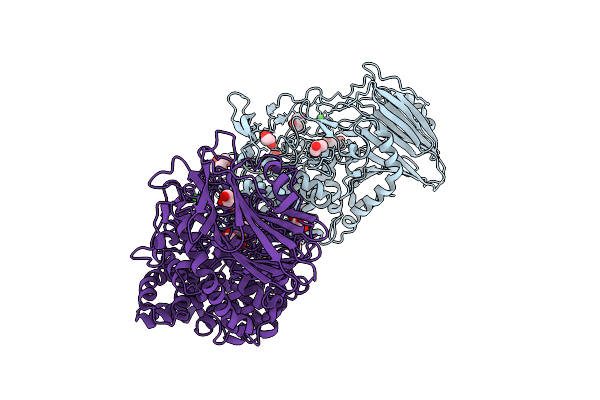 |
Crystal Structure Of Tegh116 From Thermosynechococcus Elongatus With Glucose
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: HYDROLASE Ligands: BGC, GOL, CA, TLA |
 |
 |
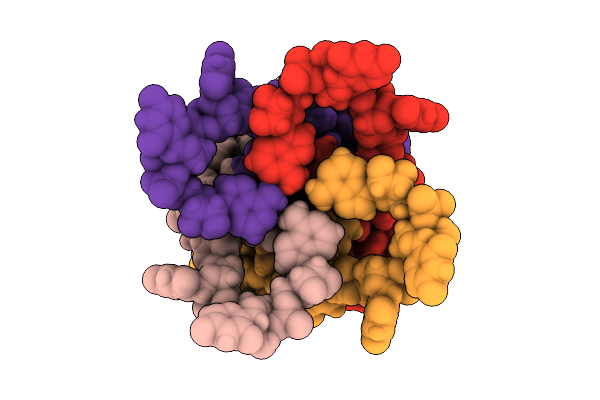
Cryo-Em Structure Of The Arabidopsis Eds1-Pad4-Adr1 Immune Complex In The Presence Of Prib-Amp
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: IMMUNE SYSTEM Ligands: AMP, RP5 |
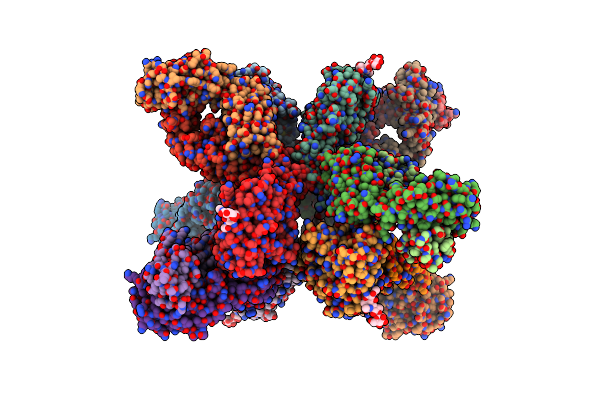 |
Human Antibody S8V1-157 In Complex With The A/American Black Duck/New Brunswick/00464/2010(H4N6) Ha Head Domain
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-10-30 Classification: Viral Protein/IMMUNE SYSTEM Ligands: MES, NAG |
 |
X-Ray Structure Of 3-Alpha-(Or 20-Beta)-Hydroxysteroid Dehydrogenase Mutant
Organism: Lactobacillaceae
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 1,4-Androstadiene-3,17- Dione
Organism: Rhodococcus electrodiphilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD, ANB |
 |
Organism: Camelus bactrianus, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: VIRAL PROTEIN |
 |
Organism: Deinococcus geothermalis
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-03-13 Classification: HYDROLASE Ligands: SO4, MN |
 |
Organism: Deinococcus geothermalis dsm 11300, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-13 Classification: HYDROLASE/DNA Ligands: MN |
 |
Organism: Influenza b virus (b/malaysia/2506/2004), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of Txgh116 From Thermoanaerobacterium Xylanolyticum With Isofagomine
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: IFM, CA, GOL, EDO |
 |
The Crystal Structure Of Trka(F589L) Kinase In Complex With N-(3-Cyclopropyl-5-((4-Methylpiperazin-1-Yl)Methyl)Phenyl)-4^6-Methyl-14-Oxo-5-Oxa-13-Aza-1(3,6)-Imidazo[1,2-B]Pyridazina-4(1,3)-Benzenacyclotetradecaphan-2-Yne-4^5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: A4U |
 |
The Crystal Structure Of Trka(G595R) Kinase In Complex With N-(3-Cyclopropyl-5-((4-Methylpiperazin-1-Yl)Methyl)Phenyl)-4^6-Methyl-14-Oxo-5-Oxa-13-Aza-1(3,6)-Imidazo[1,2-B]Pyridazina-4(1,3)-Benzenacyclotetradecaphan-2-Yne-4^5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: A4U |
 |
The Crystal Structure Of Trka Kinase In Complex With 4^6-Methyl-N-(3-(4-Methyl-1H-Imidazol-1-Yl)-5-(Trifluoromethyl)Phenyl)-14-Oxo-5-Oxa-13-Aza-1(3,6)-Imidazo[1,2-B]Pyridazina-4(1,3)-Benzenacyclotetradecaphan-2-Yne-4^5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: A0X |
 |
The Crystal Structure Of Trka Kinase In Complex With 4^6-Methyl-N-(3-(4-Methyl-1H-Imidazol-1-Yl)-5-(Trifluoromethyl)Phenyl)-11-Oxo-5-Oxa-10,14-Diaza-1(3,6)-Imidazo[1,2-B]Pyridazina-4(1,3)-Benzenacyclotetradecaphan-2-Yne-4^5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: A6X |
 |
Organism: Lentilactobacillus kefiri
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 1,4-Androstadiene-3,17- Dione
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: FAD, ANB |
 |
Crystal Structure Of Scfv Against The Receptor Binding Domine Of Sars-Cov-2 S-Spike Protein
Organism: Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-08-23 Classification: IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike Protein In Complex With Double Nabs 8H12 And 3E2 (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |

