Search Count: 84
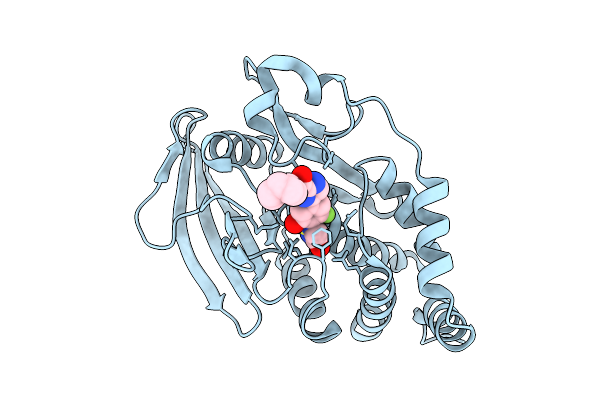 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM Ligands: A1EQR |
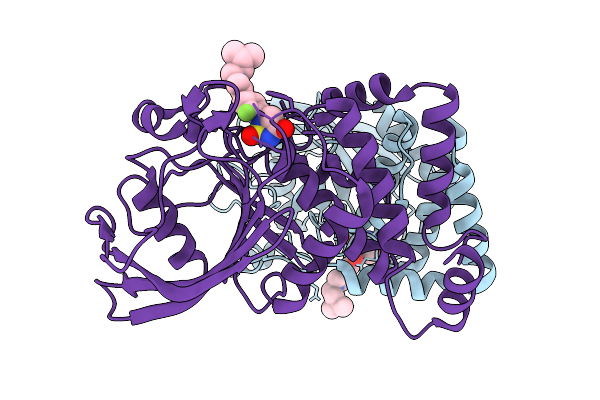 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM Ligands: A1ETL |
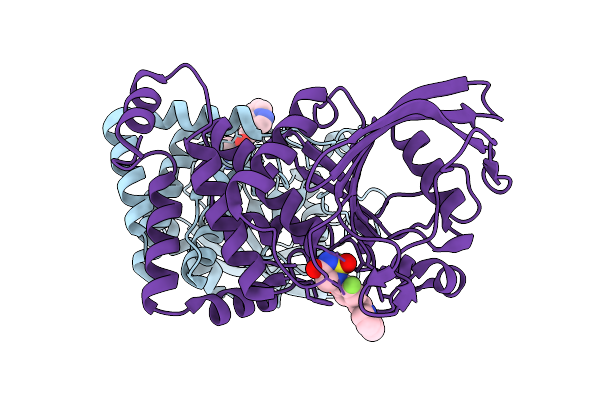 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM Ligands: A1ETK |
 |
Crystal Structure Of The S. Cerevisiae Eif2Beta N-Terminal Tail Bound To The C-Terminal Domain Of Eif5
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSLATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: PROTEIN FIBRIL |
 |
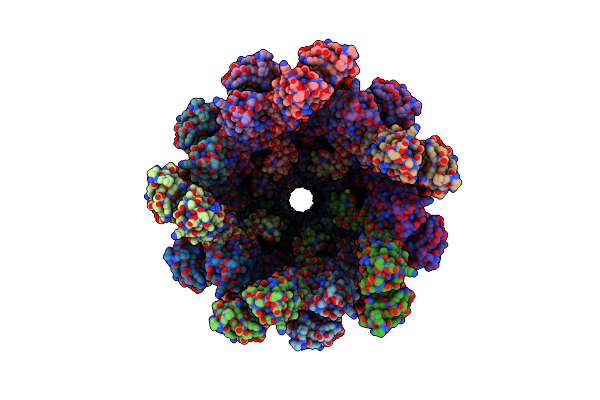
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: ANTIMICROBIAL PROTEIN Ligands: NAG |
 |
Organism: Bacillus amyloliquefaciens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: PROTEIN FIBRIL |
 |
Organism: Bacillus amyloliquefaciens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: PROTEIN TRANSPORT Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: PROTEIN TRANSPORT Ligands: NAG, CLR |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: PROTEIN TRANSPORT Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: GAY, GOL, NO3, DMS, PEG, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-06-01 Classification: TRANSFERASE/INHIBITOR Ligands: GI5, GOL, PEG |
 |
Organism: Streptomyces sp. snm55
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-12-22 Classification: HYDROLASE Ligands: CL |
 |
Chaetomium Thermophilum Udp-Glucose Glucosyl Transferase (Uggt) Double Cysteine Mutant G177C/A786C.
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2020-10-28 Classification: PROTEIN BINDING |
 |
Crystal Structure Of 2, 3-Dihydroxybenzoic Acid Decarboxylase From Fusarium Oxysporum
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-07-15 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, GOL |
 |
Crystal Structure Of 2, 3-Dihydroxybenzoic Acid Decarboxylase From Fusarium Oxysporum In Complex With Catechol
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2020-07-15 Classification: BIOSYNTHETIC PROTEIN Ligands: CAQ, ZN |
 |
Crystal Structure Of 2, 3-Dihydroxybenzoic Acid Decarboxylase From Fusarium Oxysporum In Complex With 2,5-Dhba
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-07-15 Classification: BIOSYNTHETIC PROTEIN Ligands: GTQ, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: TRANSPORT PROTEIN |

