Search Count: 71
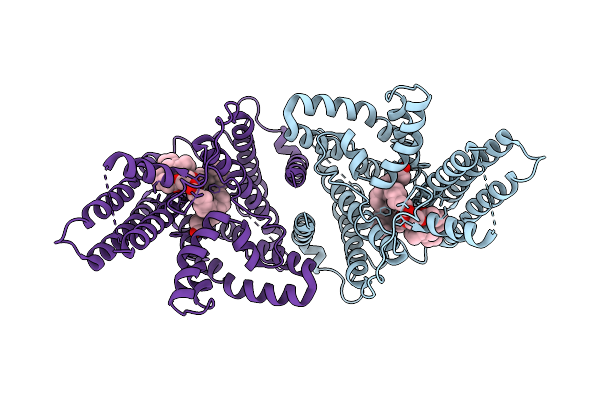 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: OLA, A1LVF |
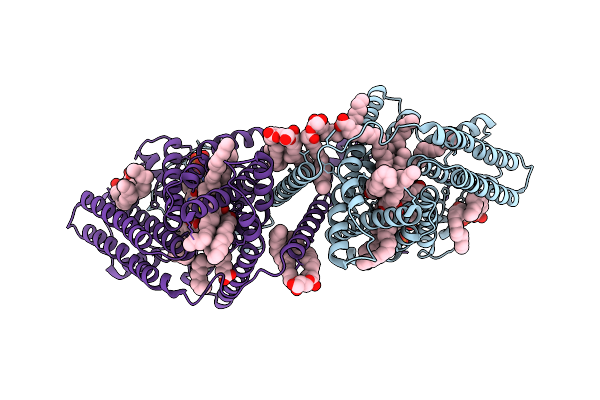 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: OLA, COA, A1LVF, LPP, A1EW5, 3VV |
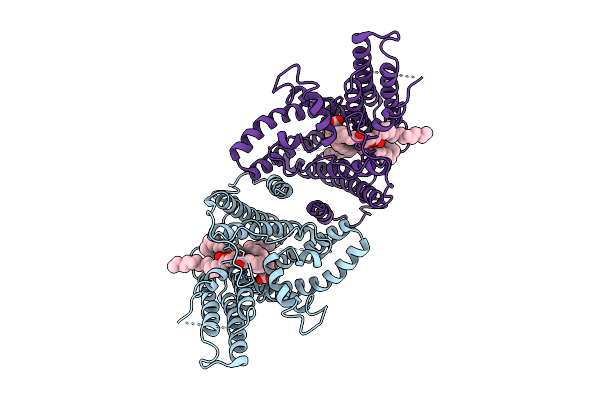 |
Structure Of The Plant Diacylglycerol O-Acyltransferase 1 In Complex With Triacylglycerol And Free Fatty Acid
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: OLA, A1LVF |
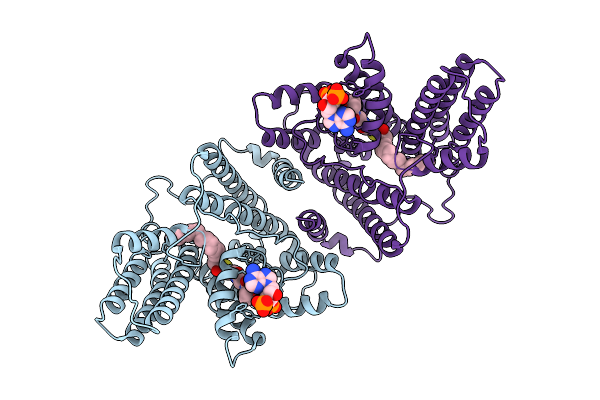 |
Structure Of The Plant Diacylglycerol O-Acyltransferase 1 In Complex With Oleoyl-Coa
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: 3VV |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: CELL CYCLE Ligands: A1JBJ |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NDP, NA |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NDP, SO4, PE8, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: A1L2M, ATP, NAG, MG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Ba.2 Rbd In Complex With Ba7535 Fab (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Delta Spike Protein In Complex With Ba7208 And Ba7125 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike (Hexapro Variant) In Complex With Nanobody W25 (Map 3, Focus Refinement On Rbd, W25 And Adjacent Ntd)
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike (Omicron Ba.1 Variant) In Complex With Nanobody W25 (Map 5, Focus Refinement On Rbd, W25 And Adjacent Ntd)
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Delta Rbd In Complex With Ba7054 And Ba7125 Fab (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Sars-Cov-2 Delta Rbd In Complex With Ba7208 And Ba7125 Fab (Local Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Spike Protein In Complex With Ba7208 Fab
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Delta Spike Protein In Complex With Ba7054 And Ba7125 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of The Ntf2L Domain Of Human G3Bp1 In Complex With The Usp10 Derived Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-01-18 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of The Ntf2L Domain Of Human G3Bp1 In Complex With The Caprin-1 Derived Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-01-18 Classification: RNA BINDING PROTEIN |
 |
Structure Of Penicillin-Binding Protein 3 (Pbp3) From Klebsiella Pneumoniae With Ligand 18G
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: JXJ |

