Search Count: 3,267
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BK4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BK3, GOL |
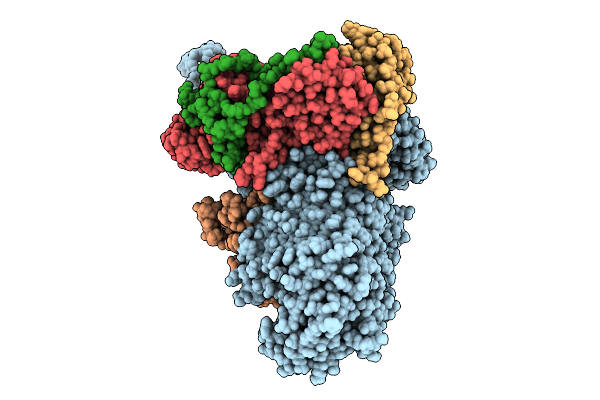 |
Beta-Barrel Assembly Machine From Escherichia Coli In An Early State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
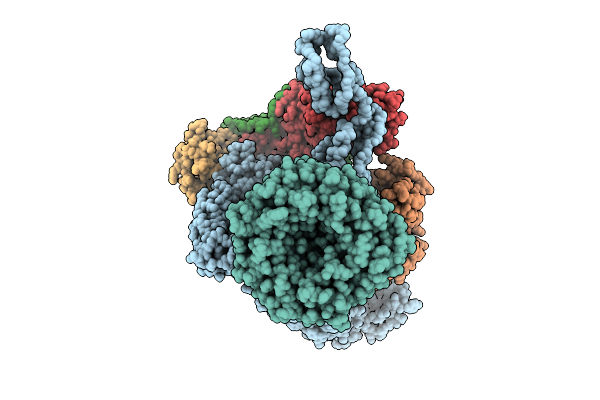 |
Beta-Barrel Assembly Machine From Escherichia Coli In A Middle State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
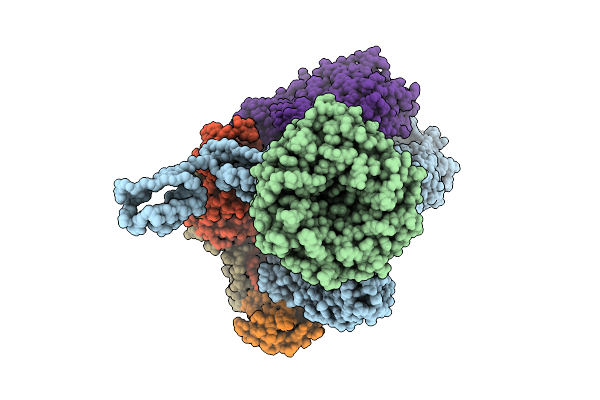 |
Beta-Barrel Assembly Machine From Escherichia Coli In A Late State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
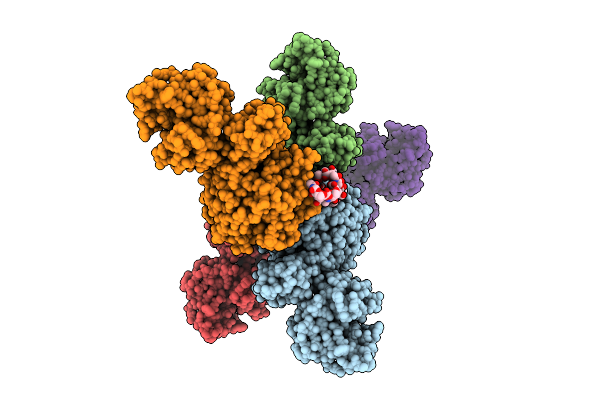 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TOXIN Ligands: A2G, A1CAY, A1CAZ |
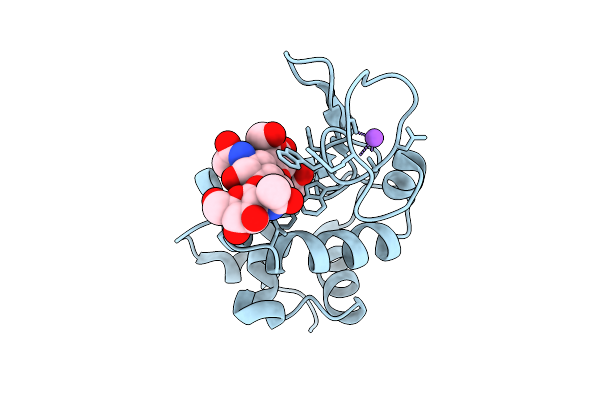 |
Structure Of Lysozyme-N,N',N"-Triacetylchitotriose Complex Determined Using Reyes For Automated Microed
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: CL, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: LIGASE Ligands: ZN, A1IW1 |
 |
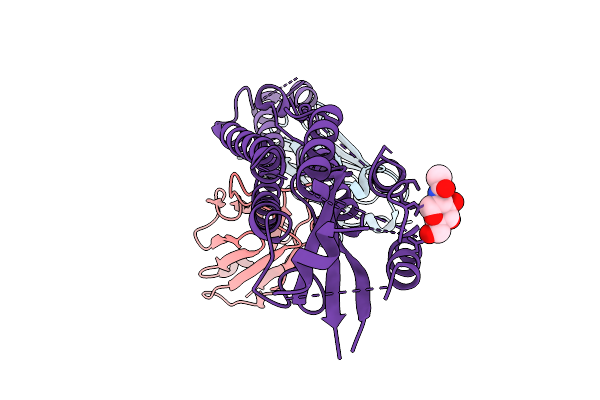
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: LIGASE Ligands: ZN, A1IW1 |
 |
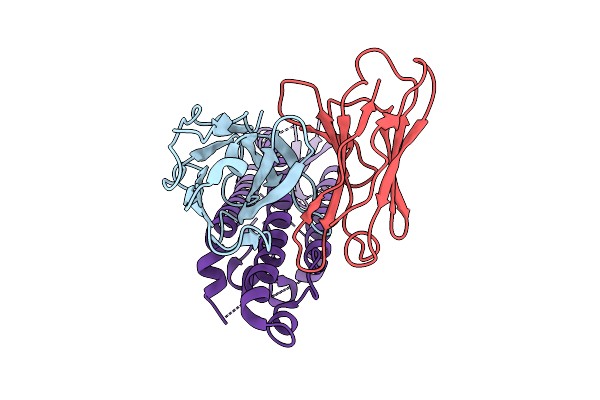
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS |
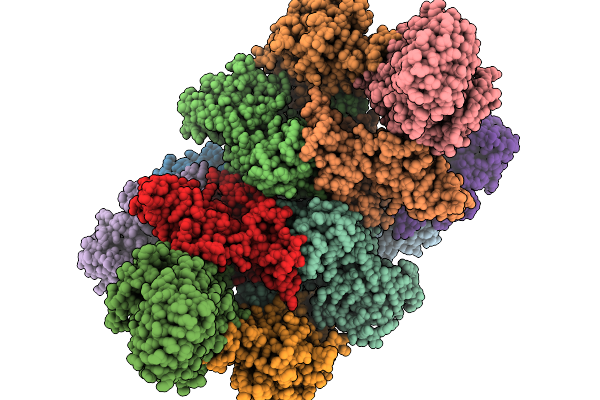 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSLATION |
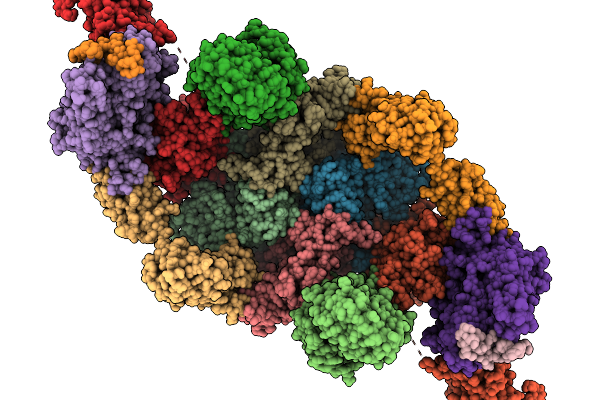 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSLATION Ligands: GTP |
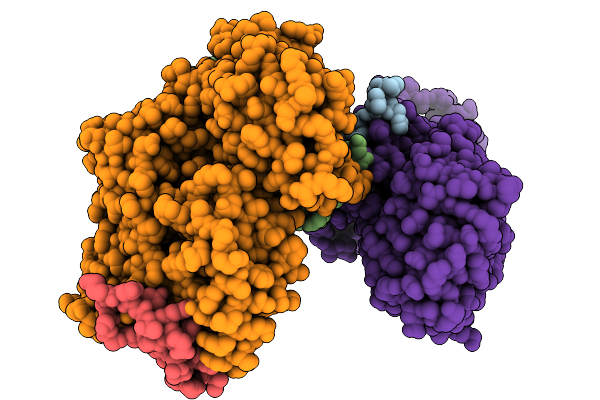 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSLATION |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN |
 |
Structure Of Yiua From Yersinia Ruckeri With Iron And Nitrilotriacetic Acid
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN Ligands: NTA, FE |
 |
Cryo-Em Structure Of The Isethionate Trap Transporter Iseqm From Oleidesulfovibrio Alaskensis With Bound Isethionate
Organism: Oleidesulfovibrio alaskensis g20, Helicobacter pylori
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: FE, A1I15, SO4 |
 |
Cryo-Em Structure Of Ternary Complex Bcl6-Crbn-Ddb1 With Bcl6-760 (Ldd, Local Refined)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: LIGASE Ligands: ZN, A1CM7 |

