Search Count: 1,136
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: RNA BINDING PROTEIN |
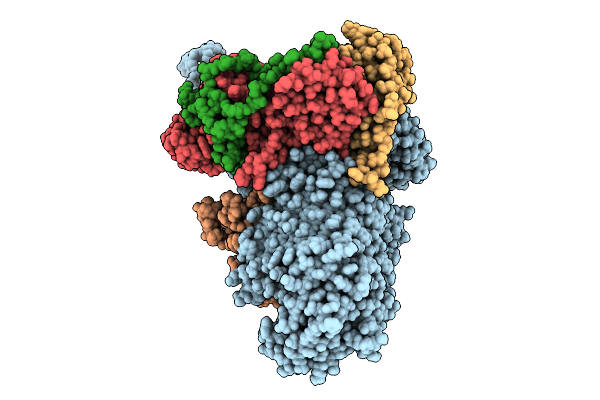 |
Beta-Barrel Assembly Machine From Escherichia Coli In An Early State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
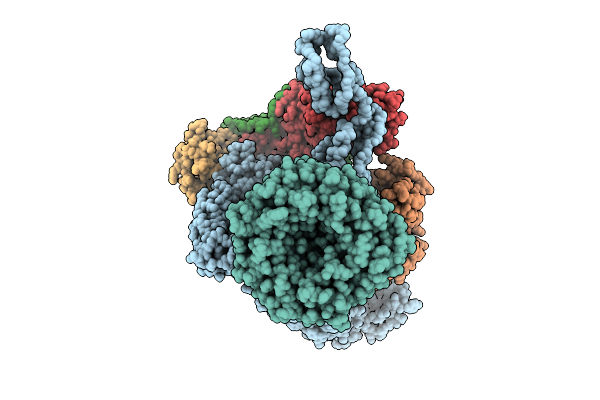 |
Beta-Barrel Assembly Machine From Escherichia Coli In A Middle State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
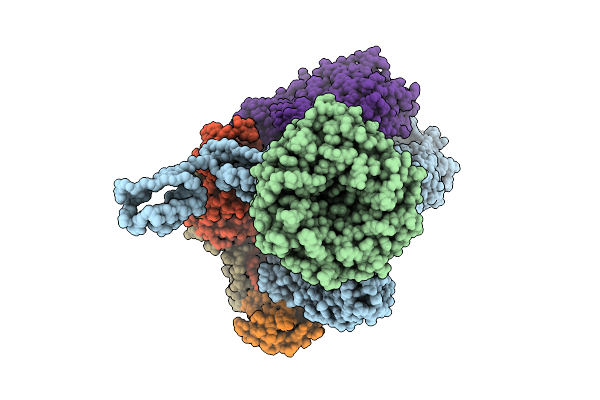 |
Beta-Barrel Assembly Machine From Escherichia Coli In A Late State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
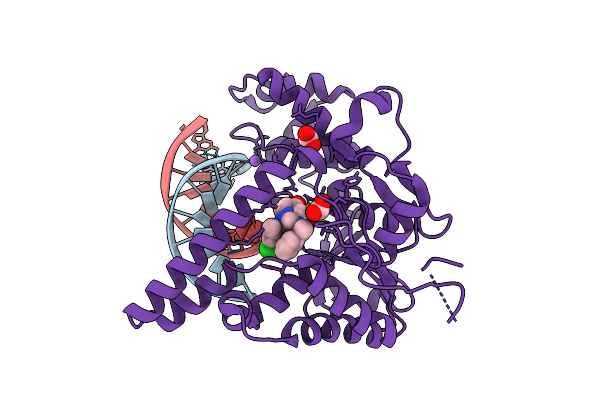 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE Ligands: MG, NA, A1JNC, FMT |
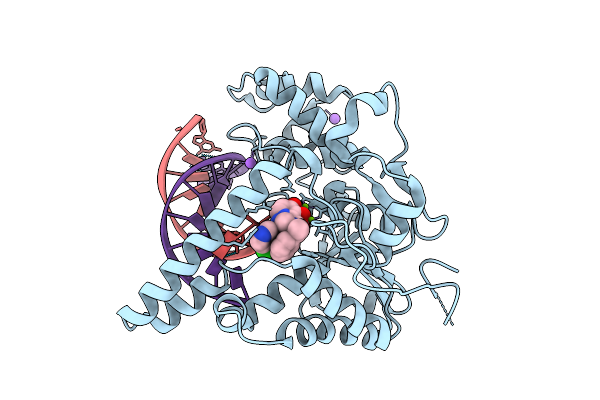 |
Crystal Structure Of Human Exonuclease 1 (Exo1) With Dna And Compound Art5537
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE Ligands: MG, A1JO0, NA |
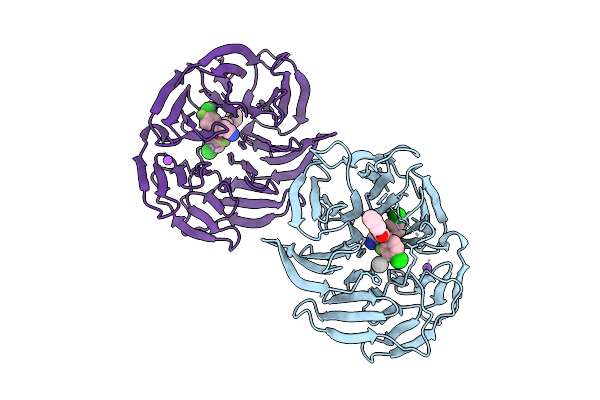 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: A1CY9, NA, UNX |
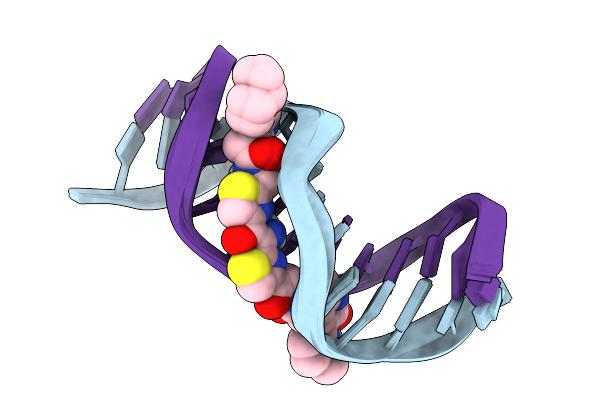 |
The Specificity And Structure Of Dna Crosslinking By A Gut Bacterial Genotoxin
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2025-11-19 Classification: DNA Ligands: A1CEC |
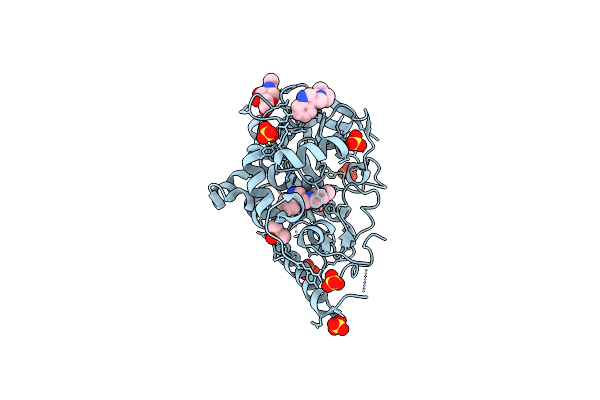 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE Ligands: SO4, NAG, DMS, LDV |
 |
Cryoem Structure Of Respiratory Syncytial Virus Polymerase In Complex With Novel Non-Nucleoside Inhibitor Compound 16
Organism: Human respiratory syncytial virus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN,TRANSFERASE/INHIBITOR |
 |
Cryoem Map Of Respiratory Syncytial Virus Polymerase With Non-Nucleoside Inhibitor Compound 21
Organism: Human respiratory syncytial virus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: MG, EDO, A1JD4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: A1JED, TEW, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSLOCASE Ligands: A1CHO, EDO, GOL |
 |
X-Ray Crystal Structure Of Kohinoor Reversibly Switchable Fluorescent Protein
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: FLUORESCENT PROTEIN Ligands: PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: A1CE0, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: A1CI0, EDO, UNX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: PROTEIN BINDING Ligands: ACT, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: A1A3I |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: A1A3J |

