Search Count: 66
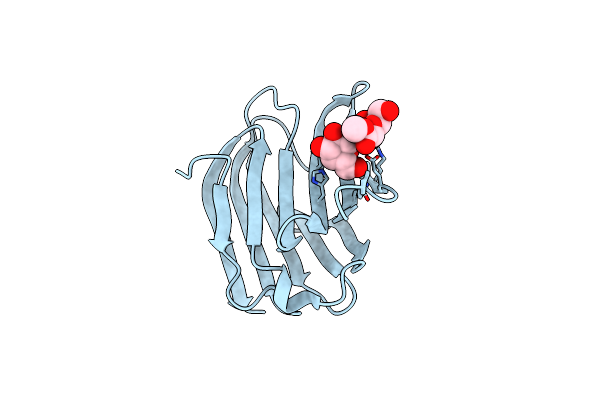 |
Galectin-3 In Complex With Methyl 2,6-Anhydro-3-Deoxy-3-S-(B-D-Galactopyranosyl)-3-Thio-D-Glycero-L-Altro-Heptonate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-09-13 Classification: SUGAR BINDING PROTEIN Ligands: CL, YIO, VPH |
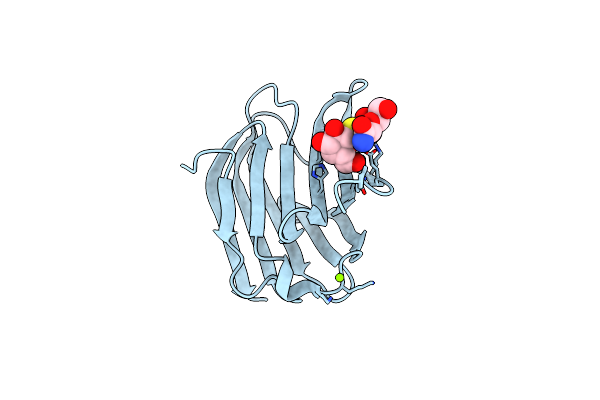 |
Galectin-3 In Complex With 2,6-Anhydro-3-Deoxy-3-S-(Beta-D-Galactopyranosyl)-3-Thio-D-Glycero-L-Altro-Heptonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: SUGAR BINDING PROTEIN Ligands: YIO, VPL, MG |
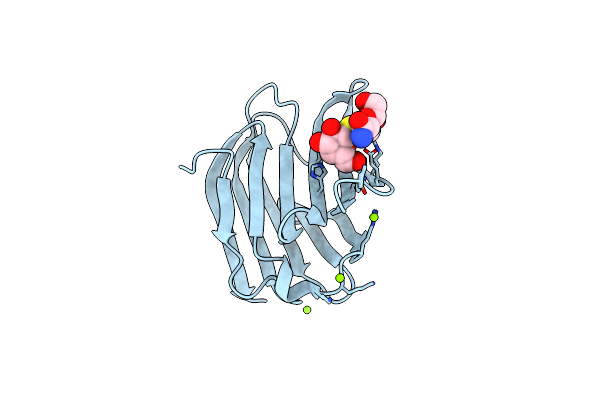 |
Galectin-3 In Complex With 2,6-Anhydro-3-Deoxy-3-S-(Beta-D-Galactopyranosyl)-3-Thio-D-Glycero-D-Galacto-Heptonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: SUGAR BINDING PROTEIN Ligands: VPQ, YIO, MG |
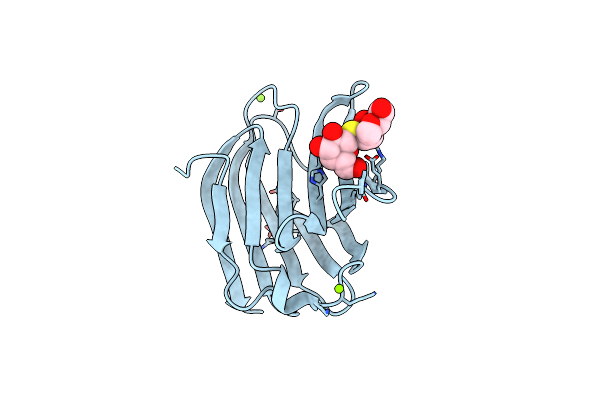 |
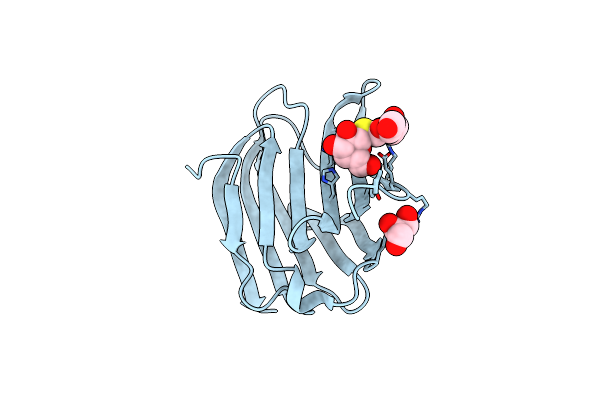
Galectin-3 In Complex With 2,6-Anhydro-5-S-(Beta-D-Galactopyranosyl)-5-Thio-D-Altritol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: SUGAR BINDING PROTEIN Ligands: MG, AH2, YIO |
 |
Galectin-3 Carbohydrate Recognition Domain In Complex With Thiodigalactoside At 1.8 Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: SUGAR BINDING PROTEIN Ligands: CL, GOL |
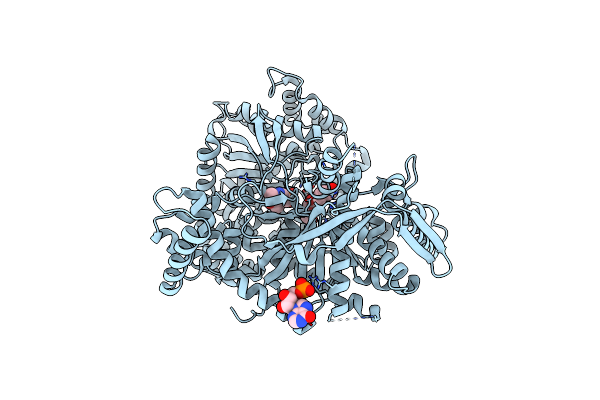 |
The Binding Of P-Coumaroyl Glucose To Glycogen Phosphorylase Reveals The Relationship Between Structural Data And Effects On Cell Metabolome
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: PLP, IMP, VKK |
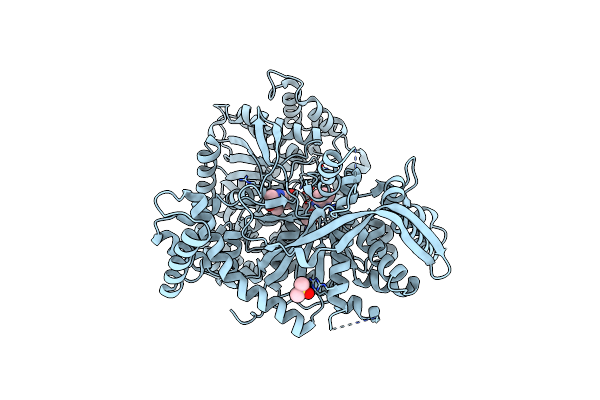 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-02-19 Classification: TRANSFERASE Ligands: KUQ, DMS, PLP |
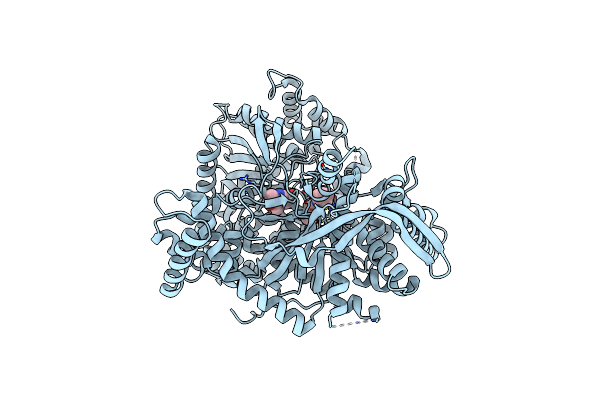 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2020-02-19 Classification: TRANSFERASE Ligands: KVW, PLP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-02-19 Classification: TRANSFERASE Ligands: KV5, PLP, IMP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2020-02-19 Classification: TRANSFERASE Ligands: KVE, PLP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-02-19 Classification: TRANSFERASE Ligands: KVH, PLP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2020-02-19 Classification: TRANSFERASE Ligands: KVQ, PLP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2020-02-19 Classification: TRANSFERASE Ligands: KVN, PLP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-06-26 Classification: TRANSFERASE Ligands: DMS, HT8, PLP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-06-26 Classification: TRANSFERASE Ligands: HTW, PLP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2019-06-26 Classification: TRANSFERASE Ligands: HTE, PLP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: PLP, B0W, IMP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: IMP, PLP, B0Z |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: B1H, PLP, IMP |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: B1K, DMS, PLP |

