Search Count: 25
 |
Stmpr1, Stenotrophomonas Maltophilia Protease 1, 36 Kda Alkine Serine Protease
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: GOL, CA, SO4 |
 |
Stmpr1, Stenotrophomonas Maltophilia Protease 1, 36 Kda Alkine Serine Protease In Complex With Bortezomib
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: BO2, GOL, SO4, CA |
 |
Stmpr1, Stenotrophomonas Maltophilia Protease 1, 36 Kda Alkine Serine Protease In Complex With Pmsf
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: GOL, PMS, SO4, CA |
 |
Stmpr1, Stenotrophomonas Maltophilia Protease 1, 36 Kda Alkine Serine Protease In Complex With Chymostatin
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: CA, SO4, GOL, A1I1B |
 |
Stmpr1, Stenotrophomonas Maltophilia Protease 1, 36 Kda Alkine Serine Protease In Complex With Leupeptin
Organism: Stenotrophomonas maltophilia, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: CA, SO4, GOL |
 |
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:3.43 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, ACT, SO4 |
 |
Organism: Blautia obeum
Method: X-RAY DIFFRACTION Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: SAM, BTB, Q46, SF4, DTB, FE, CL |
 |
Organism: Veillonella parvula
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: Q46, SAM, SF4, DTB, NA |
 |
Crystal Structure Of A Putative Nuclease With Anti-Cas9 Activity From An Uncultured Clostridia Bacterium
Organism: Clostridia bacterium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-09-22 Classification: UNKNOWN FUNCTION Ligands: NO3, MPD |
 |
The N-Terminus Of Varicella-Zoster Virus Glycoprotein B Has A Functional Role In Fusion.
Organism: Human herpesvirus 3
Method: ELECTRON MICROSCOPY Release Date: 2021-01-20 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Varicella-zoster virus (strain oka vaccine)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-07-15 Classification: VIRAL PROTEIN Ligands: NAG |
 |
A 2.8 Angstrom Cryo-Em Structure Of A Glycoprotein B-Neutralizing Antibody Complex Reveals A Critical Domain For Herpesvirus Fusion Initiation
Organism: Homo sapiens, Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Halothece sp. pcc 7418
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2018-04-11 Classification: STRUCTURAL PROTEIN |
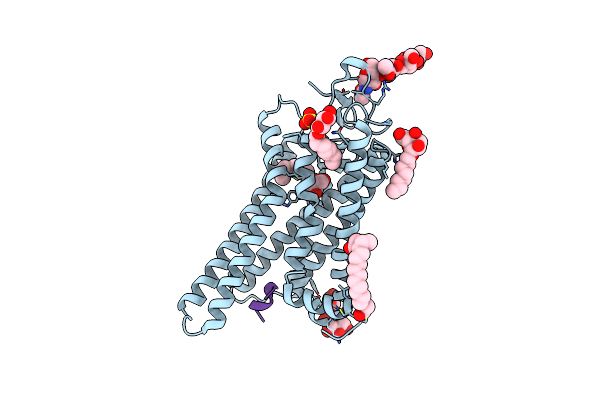 |
Crystal Structure Of The Active G-Protein-Coupled Receptor Opsin In Complex With The Finger-Loop Peptide Derived From The Full-Length Arrestin-1
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2014-09-17 Classification: SIGNALING PROTEIN Ligands: BOG, PLM, SO4, ACT |
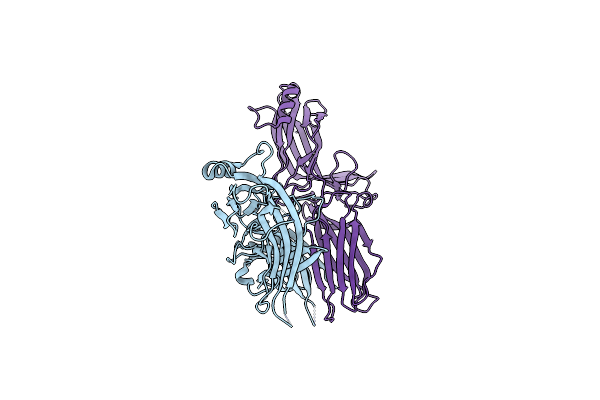 |
Crystal Structure Of C-Terminally Truncated Arrestin Reveals Mechanism Of Arrestin Activation
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-05-01 Classification: SIGNALING PROTEIN |
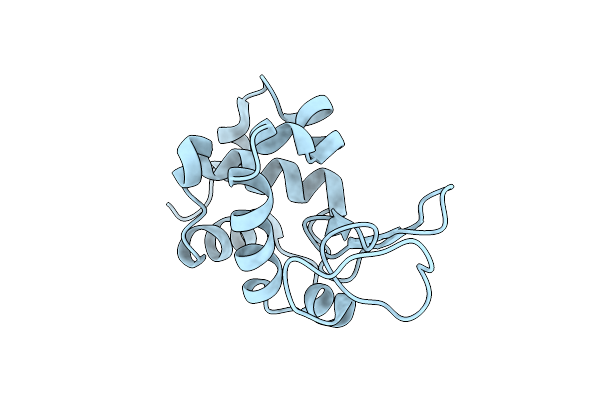 |
Crystallization And In Situ Data Collection Of Lysozyme Using The Crystal Former
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-03-30 Classification: HYDROLASE |
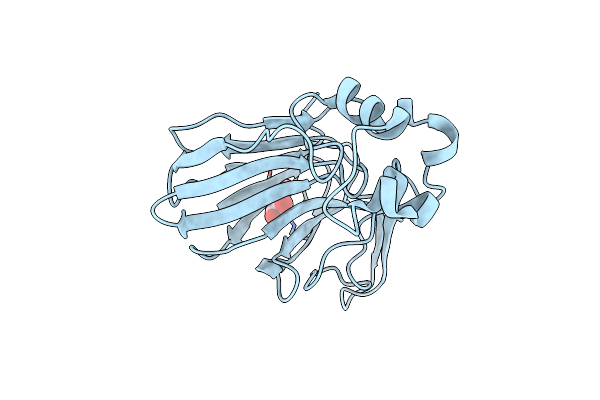 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2011-03-30 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Nostoc sp.
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2010-04-21 Classification: MEMBRANE PROTEIN |
 |
Organism: Nostoc sp.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-04-21 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Analysis Of N-Acetylneuraminate Lyase From Haemophilus Influenzae: Crystal Form I
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2000-12-20 Classification: LYASE |

