Search Count: 38
 |
Organism: Streptococcus thermophilus
Method: SOLUTION NMR Release Date: 2025-10-22 Classification: NUCLEAR PROTEIN |
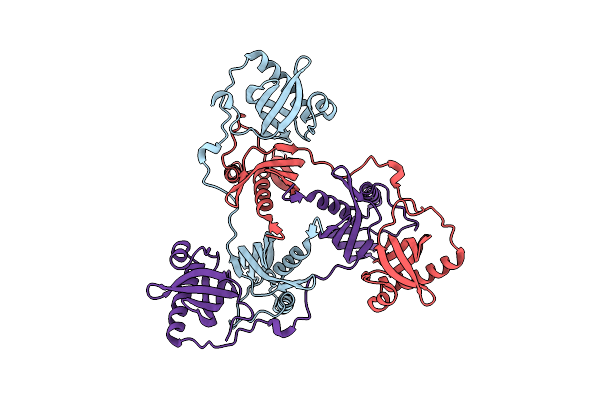 |
Crystal Structure Of A Double Mutant Of Virb8-Like Orfg Central And C-Terminal Domains Of Streptococcus Thermophilus Icest3 (Gram Positive Conjugative Type Iv Secretion System).
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-03-12 Classification: TRANSPORT PROTEIN |
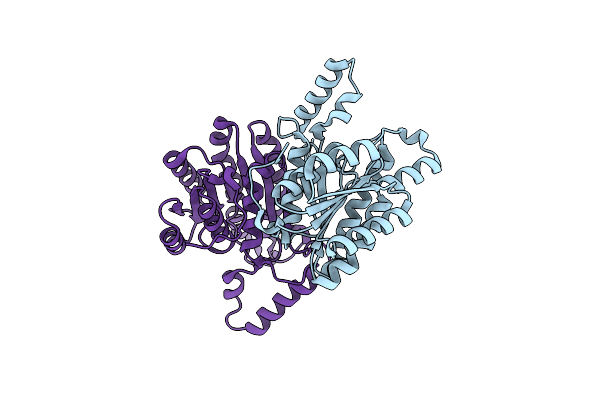 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE |
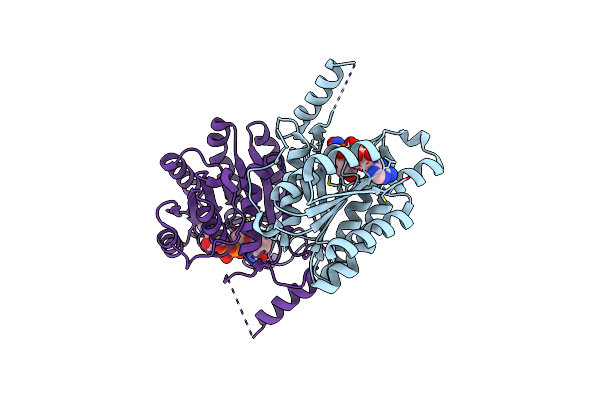 |
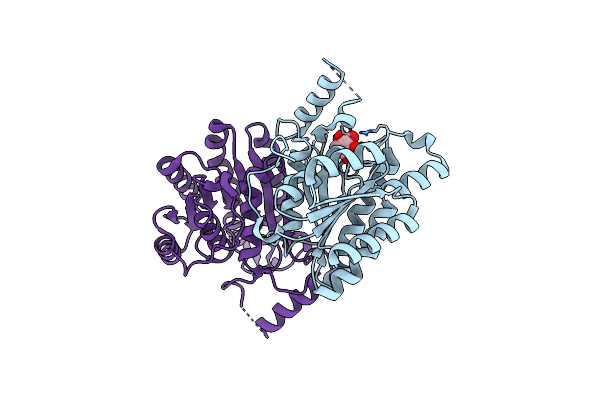
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Nad+ (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
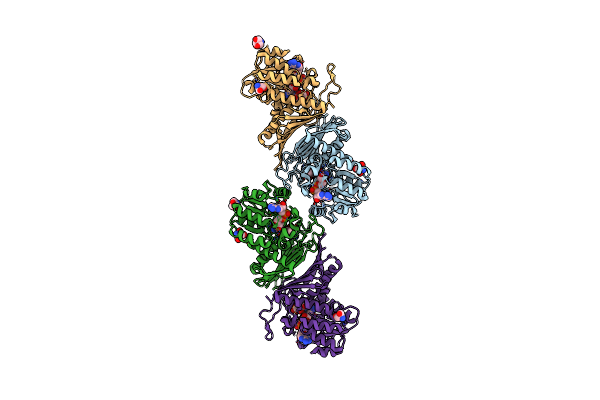
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Sulfoquinovose Substrate (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: YZT |
 |
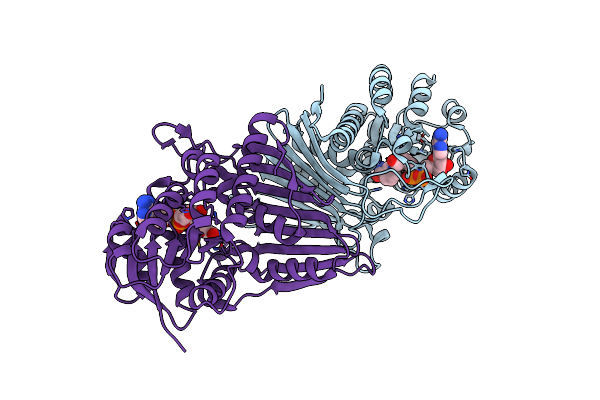
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Flavobacterium Sp. (Strain K172) In Complex With Co-Factor Nad+ And Sulfoquinovose (Sq)
Organism: Paenarthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD, R7R, TRS |
 |
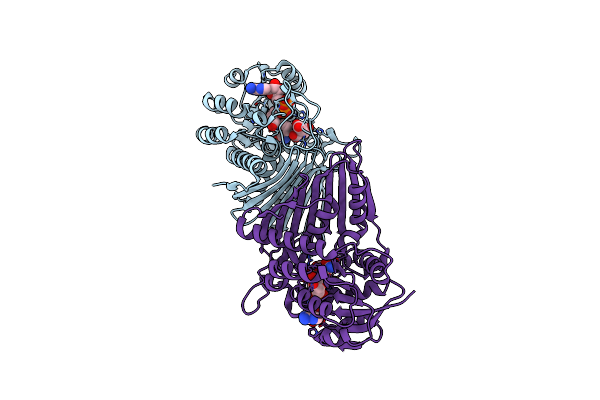
Crystal Structure Of Oxidoreductive Sulfoquinovosidase From Arthrobacter Sp. U41 (Arsqga)In Complex With Co-Factor Nad+
Organism: Arthrobacter sp. u41
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD |
 |
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Arthrobacter Sp. U41 In Complex With Nad+ Cofactor And Citrate
Organism: Arthrobacter sp. u41
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD, CIT |
 |
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Arthrobacter Sp. U41 In Complex With Nad+ And Sulfoquinovose (Sq)
Organism: Arthrobacter sp. u41
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD, R7R |
 |
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Flavobacterium Sp. (Strain K172) In Complex With Co-Factor Nad+
Organism: Paenarthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: PO4, NAD |
 |
Crystal Structure Of Class Ii Sfp Aldolase From Yersinia Aldovae (Yasqia-Zn-So4) With Bound Sulfate Ions
Organism: Yersinia aldovae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-25 Classification: LYASE Ligands: SO4, ZN, NA, BTB |
 |
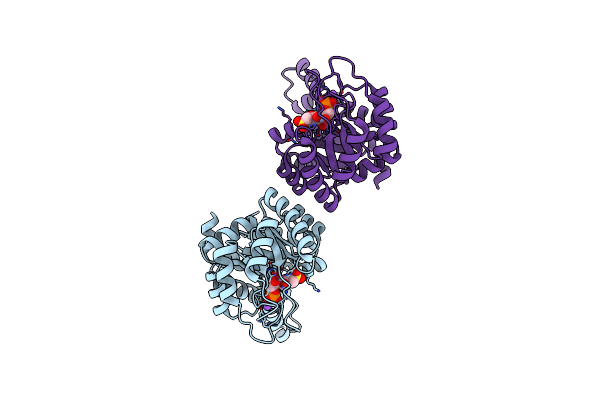
Crystal Structure Of Metal-Dependent Classii Sulfofructosephosphate Aldolase (Sfpa) From Hafnia Paralvei Hpsqia-Zn
Organism: Hafnia paralvei
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-25 Classification: LYASE Ligands: ZN |
 |
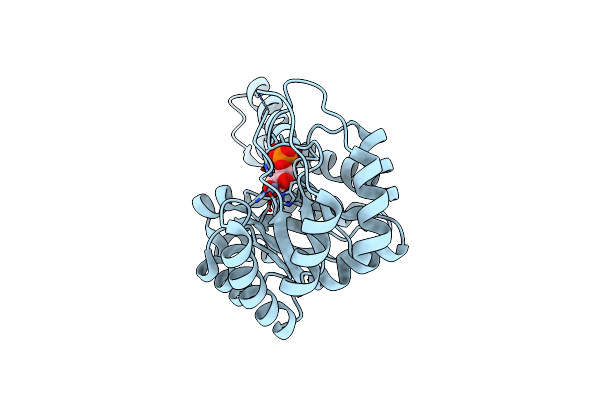
Crystal Structure Of Metal-Dependent Class Ii Sulfofructose Phosphate Aldolase From Yersinia Aldovae In Complex With Sulfofructose Phosphate (Yasqia-Zn-Sfp)
Organism: Yersinia aldovae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-10-25 Classification: LYASE |
 |
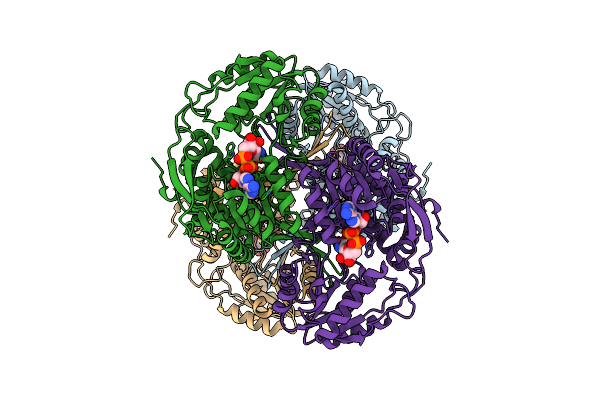
Crystal Structure Of Metal-Dependent Class Ii Sulfofructosephosphate Aldolase From Hafnia Paralvei Hpsqia-Zn In Complex With Dihydroxyacetone Phosphate (Dhap)
Organism: Hafnia paralvei
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-10-25 Classification: LYASE Ligands: ZN, 13P |
 |
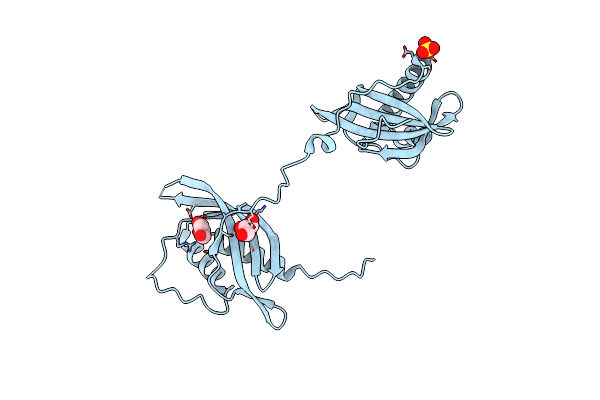
Cryo-Em Structure Of Nadh Bound Sla Dehydrogenase Rlgabd From Rhizobium Leguminosarum Bv. Trifolii Srd1565
Organism: Rhizobium leguminosarum bv. trifolii srdi565
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
Crystal Structure Of Virb8-Like Orfg Central And C-Terminal Domains Of Streptococcus Thermophilus Icest3 (Gram Positive Conjugative Type Iv Secretion System).
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-09-07 Classification: TRANSPORT PROTEIN Ligands: SO4, GOL |
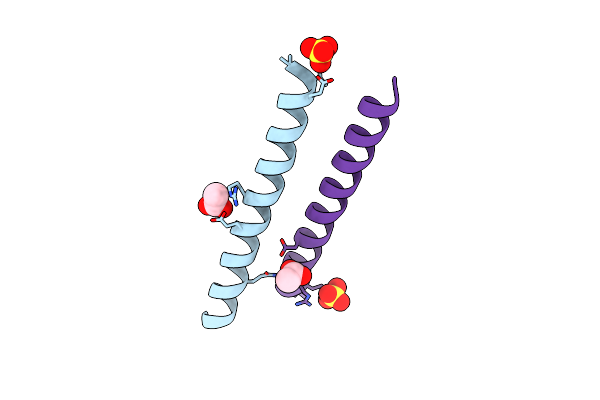 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-03-17 Classification: STRUCTURAL PROTEIN Ligands: SO4, ACT |
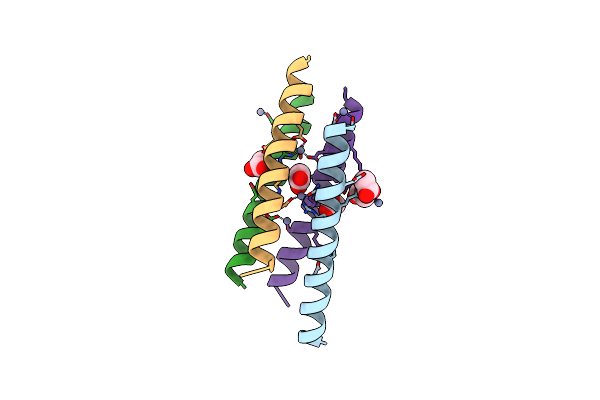 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-03-17 Classification: STRUCTURAL PROTEIN Ligands: ZN, ACT |
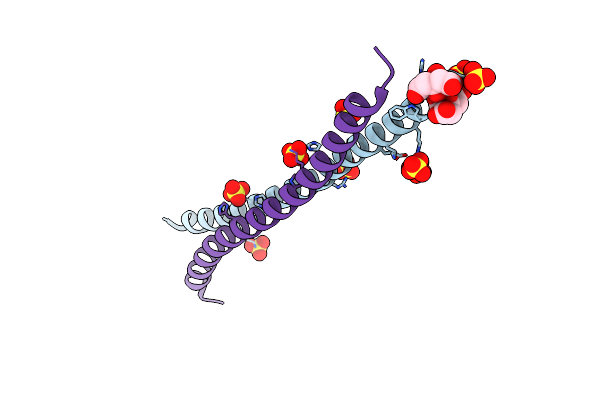 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-03-17 Classification: STRUCTURAL PROTEIN Ligands: SO4, CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-03-17 Classification: STRUCTURAL PROTEIN |

