Search Count: 24
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: A1IL0 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: A1IL7, SO4, GOL, EDO |
 |
Organism: Hepatitis b virus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2024-09-25 Classification: VIRAL PROTEIN Ligands: A1AVK |
 |
Organism: Hepatitis b virus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-09-06 Classification: VIRAL PROTEIN Ligands: YWE, EDO |
 |
Organism: Hepatitis b virus genotype a
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-05-04 Classification: VIRAL PROTEIN/INHIBITOR Ligands: 8EI |
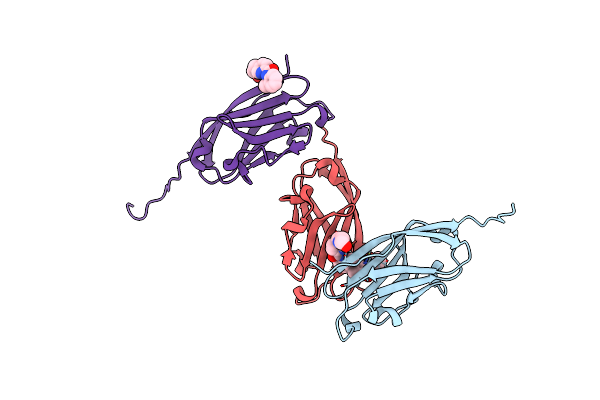 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-01-20 Classification: CELL CYCLE Ligands: R81 |
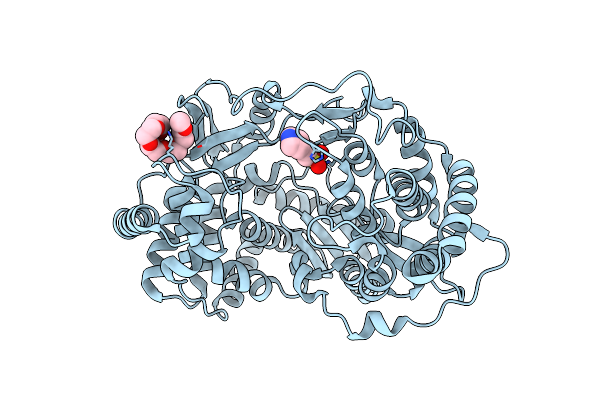 |
Apo Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With E86Q E87Q S15G C223H V321I And Delta8 Mutations
Organism: Hepatitis c virus jfh-1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-02-11 Classification: TRANSFERASE Ligands: CL, EPE, PG6 |
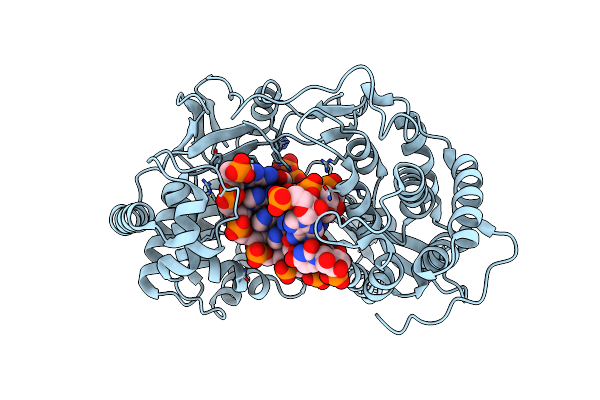 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Udp, Mn2+ And Symmetrical Primer Template 5'-Caaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, UDP |
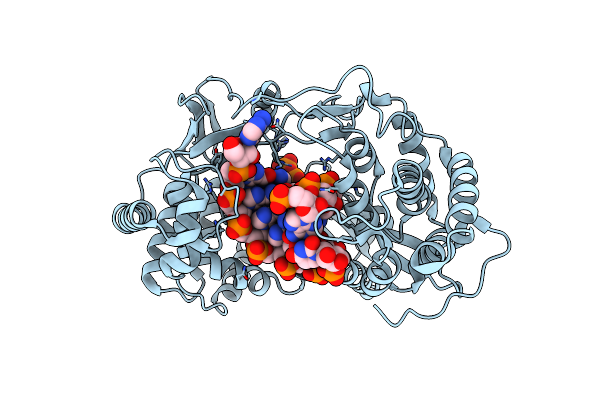 |
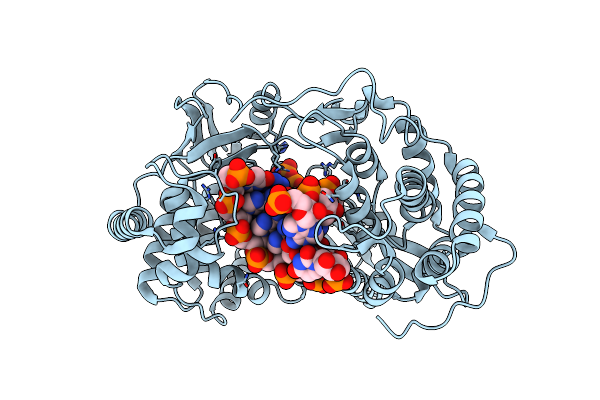
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Cdp, Mn2+ And Symmetrical Primer Template 5'-Agaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, CDP |
 |
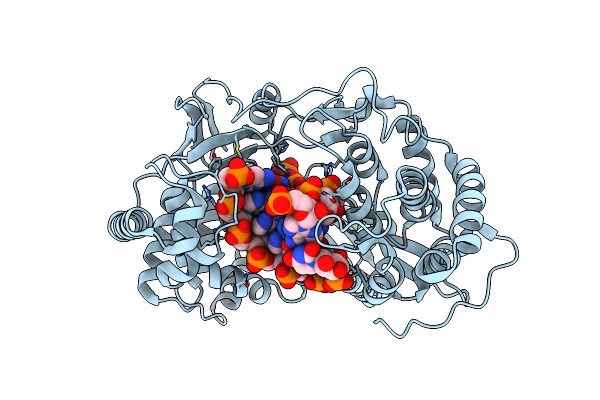
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Adp, Mn2+ And Symmetrical Primer Template 5'-Auaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, ADP, CL |
 |
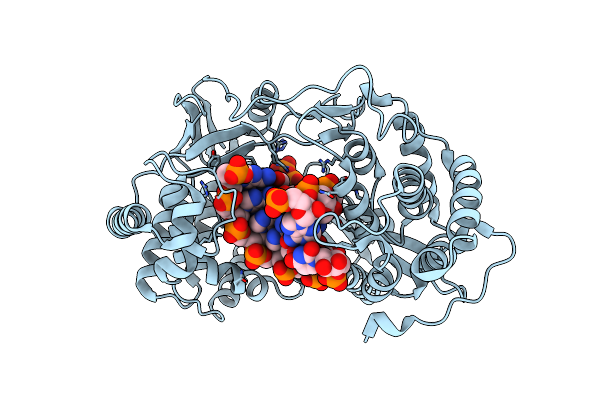
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Gdp, Mn2+ And Symmetrical Primer Template 5'-Acaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, GDP |
 |
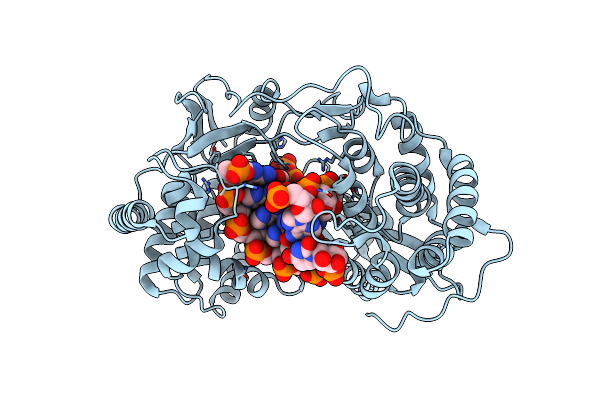
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Gs-639475, Mn2+ And Symmetrical Primer Template 5'-Caaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, 5GS |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations And Delta8 Beta Hairpin Loop Deletion In Complex With Sofosbuvir Diphosphate Gs-607596, Mn2+ And Symmetrical Primer Template 5'-Caaaauuu
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, 6GS, CL |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations In Complex With Rna Template 5'-Acgg, Rna Primer 5'-Pcc, Mn2+, And Gdp
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, GDP, B3P |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations In Complex With Rna Template 5'-Aucc, Rna Primer 5'-Pgg, Mn2+, And Adp
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, ADP, PG6, B3P, PG4, EDO |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations In Complex With Rna Template 5'-Agcc, Rna Primer 5'-Pgg, Mn2+, And Cdp
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, CDP, PG6, B3P, EDO |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations In Complex With Rna Template 5'-Uacc, Rna Primer 5'-Pgg, Mn2+, And Udp
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, UDP, PG6, B3P |
 |
Crystal Structure Of Hcv Ns5B Genotype 2A Jfh-1 Isolate With S15G E86Q E87Q C223H V321I Mutations In Complex With Rna Template 5'-Uagg, Rna Primer 5'-Pcc, Mn2+, And Udp
Organism: Hepatitis c virus jfh-1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-02-11 Classification: Transferase/RNA Ligands: MN, CL, UDP |
 |
Crystal Structure Of Hcv Polymerase Ns5B Genotype 2A Jfh-1 Isolate With The S15G, C223H, V321I Resistance Mutations Against The Guanosine Analog Gs-0938 (Psi-3529238)
Organism: Hepatitis c virus jfh-1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-09-10 Classification: Viral Protein, transferase Ligands: MES, GOL, PG6, CL |
 |
Crystal Structure Of Beta-Site App-Cleaving Enzyme 1 (Bace-Wt) Complex With N-(N-(4-Amino-3,5- Dichlorobenzyl)Carbamimidoyl)-3-(4-Methoxyphenyl)-5- Methyl-4-Isothiazolecarboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2012-10-10 Classification: HYDROLASE/INHIBITOR Ligands: 0VA, IOD |

